Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation - PubMed (original) (raw)
. 2017 Nov 2;171(4):877-889.e17.
doi: 10.1016/j.cell.2017.09.003. Epub 2017 Sep 28.
Francisca Rojas Ringeling 2, Caroline Vissers 3, Fadi Jacob 4, Michael Pokrass 3, Dennisse Jimenez-Cyrus 5, Yijing Su 6, Nam-Shik Kim 7, Yunhua Zhu 8, Lily Zheng 2, Sunghan Kim 8, Xinyuan Wang 6, Louis C Doré 9, Peng Jin 10, Sergi Regot 11, Xiaoxi Zhuang 12, Stefan Canzar 13, Chuan He 9, Guo-Li Ming 14, Hongjun Song 15
Affiliations
- PMID: 28965759
- PMCID: PMC5679435
- DOI: 10.1016/j.cell.2017.09.003
Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation
Ki-Jun Yoon et al. Cell. 2017.
Abstract
N6-methyladenosine (m6A), installed by the Mettl3/Mettl14 methyltransferase complex, is the most prevalent internal mRNA modification. Whether m6A regulates mammalian brain development is unknown. Here, we show that m6A depletion by Mettl14 knockout in embryonic mouse brains prolongs the cell cycle of radial glia cells and extends cortical neurogenesis into postnatal stages. m6A depletion by Mettl3 knockdown also leads to a prolonged cell cycle and maintenance of radial glia cells. m6A sequencing of embryonic mouse cortex reveals enrichment of mRNAs related to transcription factors, neurogenesis, the cell cycle, and neuronal differentiation, and m6A tagging promotes their decay. Further analysis uncovers previously unappreciated transcriptional prepatterning in cortical neural stem cells. m6A signaling also regulates human cortical neurogenesis in forebrain organoids. Comparison of m6A-mRNA landscapes between mouse and human cortical neurogenesis reveals enrichment of human-specific m6A tagging of transcripts related to brain-disorder risk genes. Our study identifies an epitranscriptomic mechanism in heightened transcriptional coordination during mammalian cortical neurogenesis.
Keywords: Mettl14; RNA methylation; autism; epitranscriptomics; human organoid; m(6)A; neurogenesis; radial glia cell; schizophrenia; transcriptional prepatterning.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
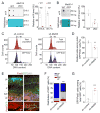
Figure 1. Nervous system Mettl14 deletion results in residual radial glia cells and ongoing neurogenesis in the postnatal mouse cortex
(A–C) Presence of neurogenic RGCs in P5 Nestin-Cre;Mettl14f/f cKO cortices. Shown are sample confocal images (A, B) and quantifications (C). Regions in white boxes are shown at a higher magnification. Scale bars: 500 μm (A, top panel), 50 μm (A, bottom panel), 100 μm (B). Values in (C) represent mean ± SEM (n = 4–7; ***: P < 0.001; *: P < 0.05; unpaired Student’s t-test). (D–E) Presence of IPCs in P5 cKO cortices. Shown are sample confocal images (D; scale bars: 100 μm) and quantification (E). Values represent mean ± SEM (n = 6; ***: P < 0.001; **: P < 0.01; unpaired Student’s t-test). (F–G) Ongoing neurogenesis in P5 cKO cortices. P5 pups were injected with EdU and analyzed 48 hr later. Shown in (F) are sample confocal images of the ventricular side of the primary somatosensory cortex. Arrows indicate Pax6+EdU+ cells (top) and Tbr2+TuJ1+EdU+ cells (bottom). Scale bars: 100 μm. Quantification of EdU+ cells with different markers is shown in (G). Values represent mean ± SEM (n = 6; ***: P < 0.001; unpaired Student’s t-test). (H–K) Reduced production of upper-layer neurons and astrocytes in cKO cortices. Pregnant mice were injected with EdU at E15.5 and analyzed at P5. Shown are sample confocal images (H, J; scale bars: 100 μm) and quantification (I, K). Values represent mean ± SEM (n = 6; ***: P < 0.001; unpaired Student’s t-test). See also Figure S1
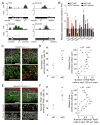
Figure 2. Mettl14−/− RGCs and NPCs exhibit prolonged cell cycle progression
(A–B) Abnormal INM of RGCs in Mettl14 cKO cortices. Pregnant mice were injected with EdU at E17.5 and analyzed 0.5 or 6 hr later. Shown are sample confocal images (A; scale bars: 50 μm) and quantification of the distance from Pax6+EdU+ nuclei to the ventricular surface (B). Values for the percentages of nuclei in each 20 μm bin represent mean ± SEM (n = 4; ***: P < 0.001; **: P < 0.01; *: P < 0.05; unpaired Student’s t-test). (C–D) Delayed S to M phase transition of RGCs in Mettl14 cKO mice. Pregnant mice were injected with EdU at E17.5 and analyzed 2 hr later. Shown in (C) are sample confocal images. Arrowheads point to Pax6+pH3+EdU+ cells and arrows point to Pax6+pH3+EdU− cells. Scale bar: 50 μm. Shown in (D) is the quantification of the percentage of Pax6+pH3+EdU+ cells, representing cells proceeded from S to M phase during the 2 hr chase, among total Pax6+pH3+ cells. Values represent mean ± SEM (n = 5 for WT and n = 8 for cKO; ***: P < 0.001; unpaired Student’s t-test). (E–F) Delayed cell cycle exit of neural progenitors in Mettl14 cKO mice. Pregnant mice were injected with EdU at E17.5 and analyzed 24 hr later. Shown in (E) are sample confocal images. Arrowheads point to Ki67−EdU+ cells and arrows point to Ki67+EdU− cells. Scale bar: 50 μm. Shown in (F) is the quantification of the percentage of Ki67−EdU+ cells, representing cells exited from cell cycle, among total EdU+ cells. Values represent mean ± SEM (n = 6; ***: P < 0.001; unpaired Student’s t-test). (G–J) Time-lapse imaging analysis of mouse NPCs showing prolonged S-G2-M phase length in the absence of Mettl14. WT and cKO mouse NPCs were electroporated with plasmid co-expressing a Cdk2 sensor (green) and the H2B-mCherry nuclear marker (red), cultured for 2 days, and imaged for 48 hr. Shown in (G) are sample time-lapse images with time stamps. Scale bars: 10 μm. Also shown are box plots of quantifications for the total cell cycle length (H; n = 38 for WT and n = 30 for cKO), G1 phase length (I; n = 20), and S-G2-M phase length (J; n = 20). Each dot represents data from one NPC (***: P < 0.001; unpaired Student’s t-test). See also Figure S2.
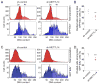
Figure 3. Mettl3 regulates cell cycle progression of NPCs and maintenance of embryonic cortical RGCs
(A) Depletion of m6A-tagging on mRNAs purified from E15.5 and E17.5 Mettl14 cKO mouse forebrain. Shown in the left panels are sample images of m6A dot blot and methylene blue staining (for loading controls). Data were normalized to the averaged levels of WT samples and quantification is shown in the right panel. Values represent mean ± SEM (n = 3; **: P < 0.01; unpaired Student’s t-test). (B) Depletion of m6A-tagging on mRNAs purified from Mettl14 cKO NPCs. Values represent mean ± SEM (n = 3; **: P < 0.01; unpaired Student’s t-test). (C–D) Flow cytometry analysis of cell cycle status of mouse NPCs. NPCs were electroporated to co-express GFP and the control shRNA, or the shRNA against Mettl3. At day 4, NPCs were pulse-labeled with EdU (10 μM) for 30 min, cultured for 9 hr, followed by EdU and DNA content (DyeCycle Violet) staining and flow cytometry analysis. Shown are sample histograms of DNA content from GFP+EdU+ cells and the total cell population (as a reference; C) and quantification (D). Values in (D) represent mean ± SEM (n = 4; **: P < 0.01; unpaired Student’s t-test). (E–G) Embryonic mouse cortices were electroporated in utero at E13.5 to co-express GFP and shRNA-control, or GFP and shRNA-Mettl3, and analyzed at E17.5. Shown in (E) are sample confocal images. Scale bars: 50 μm. The distribution of GFP+ cells in each zone (F) and the percentage of GFP+Pax6+ cells among total GFP+ cells (G) were quantified. VZ: ventricular zone; SVZ: subventricular zone; IZ: intermediate zone; CP: cortical plate. Values represent mean ± SEM (n = 4; ***: P < 0.001; **: P < 0.01; unpaired Student’s t-test). See also Figure S3.
Figure 4. m6A tags transcripts related to transcription factors, cell cycle, and neuronal differentiation in the embryonic mouse brain, and promotes their decay
(A) Coverage plots from m6A-seq of E13.5 mouse forebrains showing representative examples of m6A-tagged (Sox1, Emx2, and Cdk9) and non m6A-tagged (Rad17) transcripts. Top and middle panels show read coverages normalized by library sizes from m6A pulled-down and input libraries, respectively, and bottom panels show gene structures (arrows point to the direction of transcription; S.C.: stop codon). (B–C) GO analysis of m6A-tagged genes reveals enrichment for biological process terms related to transcription factors, neurogenesis, cell cycle, and stem cell differentiation. Also shown is Wikipathways gene set enrichment analysis. FDR: false discovery rate. (D) Cumulative distribution of Log2(gene expression ratios) at time 5 hr post ActD over time 0 hr for m6A tagged genes (purple line) and non-m6A tagged genes (black line) for WT and Mettl14 cKO NPCs. D = value of Kolmogorov Smirnov test statistic corresponding to maximum difference between methylated and non-methylated distributions. (E) Cumulative distribution of log2(fold change in ratios of gene expression) at 5 hr post ActD treatment over time 0 hr upon Mettl14 deletion. Top panel shows cumulative distribution for non-targets (black line) and transcripts with 1–1.9 m6A sites on average (bright red line), or 2 or more sites on average (dark red line). Bottom panel shows cumulative distribution for non-targets and m6A-tagged transcripts with 1–2, 2–3, 3–4, and 4 or more sites on average. (F) Summary of half-life of a group of transcripts in WT and Mettl14 cKO NPCs. Values represent mean ± SEM (n = 4; ***: P < 0.001; **: P < 0.01; unpaired Student’s t-test See also Figure S4, and Table S2, S3 and S4.
Figure 5. Post-transcriptional regulation of pre-patterning gene levels and protein production by m6A signaling in cortical neural stem cells
(A) Coverage plots from m6A-seq of E13.5 mouse forebrains showing representative examples of m6A-tagged IPC (Tbr2 and Neurog2) and neuronal (Neurod1 and Neurod2) genes. (B) Q-PCR analysis of total mRNA and 4sU-purified nascent mRNA from WT and Mettl14 cKO NPCs. All Ct values were first normalized to Gapdh control (not m6A-tagged), which were similar in both WT and cKO NPCs. The ratio (cKO over WT) was calculated for each experiment and values represent mean ± SEM (n = 3 cultures; ***: P < 0.001; **: P < 0.01; *: P < 0.05; unpaired Student’s t-test). (C–F) Precocious expression of Tbr2 and Neurod1 proteins in RGCs in E17.5 Mettl14 cKO mice in vivo. Shown are sample confocal images (C, E; scale bars: 50 μm) and quantifications of the percentage of Tbr2+Pax6+ cells (D), or Neurod1+Pax6+ cells (F), among total Pax6+ cells (left panels, n = 6) and the density distribution of Tbr2+Pax6+ (D), or Neurod1+Pax6+ cells (F), from the ventricular surface (right panels, n = 4). Arrows indicate Tbr2+Pax6+ (C) or Neurod1+Pax6+ cells (E). Values in (D, F) represent mean ± SEM (***: P < 0.001; **: P < 0.01; *: P < 0.05; unpaired Student’s t-test). See also Figure S5.
Figure 6. METTL14 regulates cell cycle progression of human NPCs
(A–B) Flow cytometry analysis of cell cycle progression of hNPCs with METTL14 KD. Human NPCs were electroporated to co-express GFP and shRNA-control, or shRNA-METTL14. After 4 days, hNPCs were pulse-labeled with EdU (10 μM) for 30 min, incubated for 14 hr, followed by EdU and DNA content (DyeCycle Violet) staining and flow cytometry analysis, similarly as in Figure 3C–D. Values represent mean ± SEM (n = 4; **: P < 0.01; unpaired Student’s t-test). (C–D) Flow cytometry analysis of cell cycle progression with METTL14 KD in human forebrain organoids. Day 45 forebrain organoids were electroporated to co-express GFP and shRNA-control, or shRNA-METTL14. After 7 days, forebrain organoids were pulse-labeled with EdU (10 μM) for 1 hr, cultured further for 14 hr, followed by dissociation and analysis similarly as in Figure 3C–D. Values represent mean ± SEM (n = 4; ***: P < 0.001; unpaired Student’s t-test). See also Figure S6.
Figure 7. Conserved and unique features of m6A mRNA methylation in human forebrain organoids, human fetal brain and embryonic mouse forebrain
(A) Representative plots of two m6A-tagged transcripts in day 47 human forebrain organoids and PCW11 human fetal brain, but not in mouse E13.5 forebrain. (B) Venn diagram showing shared m6A-tagged transcripts between day 47 human forebrain organoids and PCW11 fetal human brain. (C) GO and disease ontology analyses of shared m6A-tagged genes in day 47 human forebrain organoids and PCW11 human fetal brain. (D) Venn diagram showing shared and unique m6A-tagged transcripts among mouse E13.5 forebrain, day 47 human forebrain cortex, and PCW11 fetal human brain. Only ortholog genes expressed in all three samples were used for analysis. (E–F) Disease ontology analysis of transcripts uniquely m6A-tagged in human shows enrichment for neurodevelopmental diseases, whereas disease ontology analysis of commonly m6A-tagged transcripts showed enrichment for oncogenic processes. See also Figure S7, and Table S2 and S5.
Comment in
- Epimetronomics: m6A Marks the Tempo of Corticogenesis.
Boles NC, Temple S. Boles NC, et al. Neuron. 2017 Nov 15;96(4):718-720. doi: 10.1016/j.neuron.2017.11.002. Neuron. 2017. PMID: 29144970 - Layered-up regulation in the developing brain.
Sweatt JD. Sweatt JD. Nature. 2017 Nov 23;551(7681):448-449. doi: 10.1038/d41586-017-07269-7. Nature. 2017. PMID: 29168827 Free PMC article.
Similar articles
- m6A Regulates Neurogenesis and Neuronal Development by Modulating Histone Methyltransferase Ezh2.
Chen J, Zhang YC, Huang C, Shen H, Sun B, Cheng X, Zhang YJ, Yang YG, Shu Q, Yang Y, Li X. Chen J, et al. Genomics Proteomics Bioinformatics. 2019 Apr;17(2):154-168. doi: 10.1016/j.gpb.2018.12.007. Epub 2019 May 30. Genomics Proteomics Bioinformatics. 2019. PMID: 31154015 Free PMC article. - N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications.
Wang Y, Li Y, Yue M, Wang J, Kumar S, Wechsler-Reya RJ, Zhang Z, Ogawa Y, Kellis M, Duester G, Zhao JC. Wang Y, et al. Nat Neurosci. 2018 Feb;21(2):195-206. doi: 10.1038/s41593-017-0057-1. Epub 2018 Jan 15. Nat Neurosci. 2018. PMID: 29335608 Free PMC article. - Ythdf2-mediated m6A mRNA clearance modulates neural development in mice.
Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z, Perez SP, Suganthan R, He C, Bjørås M, Klungland A. Li M, et al. Genome Biol. 2018 May 31;19(1):69. doi: 10.1186/s13059-018-1436-y. Genome Biol. 2018. PMID: 29855337 Free PMC article. - The m6A epitranscriptome on neural development and degeneration.
Yen YP, Chen JA. Yen YP, et al. J Biomed Sci. 2021 May 27;28(1):40. doi: 10.1186/s12929-021-00734-6. J Biomed Sci. 2021. PMID: 34039354 Free PMC article. Review. - N (6)-Methyladenosine (m(6)A) Methylation in mRNA with A Dynamic and Reversible Epigenetic Modification.
Wu R, Jiang D, Wang Y, Wang X. Wu R, et al. Mol Biotechnol. 2016 Jul;58(7):450-9. doi: 10.1007/s12033-016-9947-9. Mol Biotechnol. 2016. PMID: 27179969 Review.
Cited by
- Single-cell m6A profiling in the mouse brain uncovers cell type-specific RNA methylomes and age-dependent differential methylation.
Tegowski M, Prater AK, Holley CL, Meyer KD. Tegowski M, et al. Nat Neurosci. 2024 Dec;27(12):2512-2520. doi: 10.1038/s41593-024-01768-3. Epub 2024 Sep 24. Nat Neurosci. 2024. PMID: 39317796 Free PMC article. - Liquid-liquid phase separation in diseases.
Zhang X, Yuan L, Zhang W, Zhang Y, Wu Q, Li C, Wu M, Huang Y. Zhang X, et al. MedComm (2020). 2024 Jul 13;5(7):e640. doi: 10.1002/mco2.640. eCollection 2024 Jul. MedComm (2020). 2024. PMID: 39006762 Free PMC article. Review. - Epitranscriptomics in the Heart: a Focus on m6A.
Longenecker JZ, Gilbert CJ, Golubeva VA, Martens CR, Accornero F. Longenecker JZ, et al. Curr Heart Fail Rep. 2020 Oct;17(5):205-212. doi: 10.1007/s11897-020-00473-z. Curr Heart Fail Rep. 2020. PMID: 32813261 Free PMC article. Review. - Recent advances in functional annotation and prediction of the epitranscriptome.
Zhang SY, Zhang SW, Zhang T, Fan XN, Meng J. Zhang SY, et al. Comput Struct Biotechnol J. 2021 May 21;19:3015-3026. doi: 10.1016/j.csbj.2021.05.030. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34136099 Free PMC article. Review. - The role of post-transcriptional modifications during development.
Hamar R, Varga M. Hamar R, et al. Biol Futur. 2023 Jun;74(1-2):45-59. doi: 10.1007/s42977-022-00142-3. Epub 2022 Dec 8. Biol Futur. 2023. PMID: 36481986 Review.
References
MeSH terms
Substances
Grants and funding
- P01 NS097206/NS/NINDS NIH HHS/United States
- R35 NS097370/NS/NINDS NIH HHS/United States
- RM1 HG008935/HG/NHGRI NIH HHS/United States
- T32 GM007814/GM/NIGMS NIH HHS/United States
- R21 MH087874/MH/NIMH NIH HHS/United States
- T32 GM007309/GM/NIGMS NIH HHS/United States
- T32 GM007445/GM/NIGMS NIH HHS/United States
- R21 NS093772/NS/NINDS NIH HHS/United States
- R33 MH087874/MH/NIMH NIH HHS/United States
- P30 DK089502/DK/NIDDK NIH HHS/United States
- R37 NS047344/NS/NINDS NIH HHS/United States
- U19 MH106434/MH/NIMH NIH HHS/United States
- R21 NS095348/NS/NINDS NIH HHS/United States
- R56 NS047344/NS/NINDS NIH HHS/United States
- R01 NS048271/NS/NINDS NIH HHS/United States
- R01 NS047344/NS/NINDS NIH HHS/United States
- R01 DA043361/DA/NIDA NIH HHS/United States
- R01 NS051630/NS/NINDS NIH HHS/United States
- U19 AI131130/AI/NIAID NIH HHS/United States
- T32 GM008752/GM/NIGMS NIH HHS/United States
- R01 MH102690/MH/NIMH NIH HHS/United States
- R01 MH105128/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous