Single-Cell Analysis of Human Pancreas Reveals Transcriptional Signatures of Aging and Somatic Mutation Patterns - PubMed (original) (raw)
Single-Cell Analysis of Human Pancreas Reveals Transcriptional Signatures of Aging and Somatic Mutation Patterns
Martin Enge et al. Cell. 2017.
Abstract
As organisms age, cells accumulate genetic and epigenetic errors that eventually lead to impaired organ function or catastrophic transformation such as cancer. Because aging reflects a stochastic process of increasing disorder, cells in an organ will be individually affected in different ways, thus rendering bulk analyses of postmitotic adult cells difficult to interpret. Here, we directly measure the effects of aging in human tissue by performing single-cell transcriptome analysis of 2,544 human pancreas cells from eight donors spanning six decades of life. We find that islet endocrine cells from older donors display increased levels of transcriptional noise and potential fate drift. By determining the mutational history of individual cells, we uncover a novel mutational signature in healthy aging endocrine cells. Our results demonstrate the feasibility of using single-cell RNA sequencing (RNA-seq) data from primary cells to derive insights into genetic and transcriptional processes that operate on aging human tissue.
Keywords: glucagon; human aging; human pancreas; insulin; islet; mutational signatures; single-cell RNA-seq; somatic variation; transcriptional instability.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
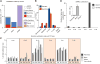
Figure 1. A Comprehensive Survey of Single Cells Sampled from Human Pancreas across Different Ages
(A) tSNE plot of 2,544 successful scRNA-seq libraries from eight donors. Each point represents one cell and points are positioned to retain pairwise distances as determined by Pearson correlation of the 500 most highly expressed genes. Cell identity is indicated by marker gene expression. (B) Fraction of cells that express the aging associated gene_CDKN2A_ (p16) in juvenile (0–6 years), young adult (21–22 years), and middle-aged (38–54 years) donors increases with age (p = 3.1E-3, n = 8, linear regression.) Bars are mean ± SEM (n = 2–3). (C) Boxplot of transcriptional noise in β-cells, plotted by age group. Higher age is associated with increased whole-transcriptome cell-to-cell variability within cell type (p = 6.67E-9, n = 384). Boxes indicate the middle quartiles, separated by median line. Whiskers indicate last values within 1.5 × the interquartile range for the box. (D) Violin plots show the ratio of Insulin–Glucagon protein staining at the sites of Insulin (INS, n = 5,801) and Glucagon (GCG, n = 3,254) RNA hybridization spots. (E) Boxplot of Log2 counts per million (CPM) of cell-atypical glucagon transcript in β cells (left), and insulin transcripts in α-cells (right), in cells from juvenile (0–6 years), young adult (21–22 years) and middle-aged (38–54) donors. Boxes indicate the middle quartiles, separated by median line. Whiskers indicate last values within 1.5 × the interquartile range for the box. See also Figure S1 and Tables S1, S2, and S3.
Figure 2. Gene Expression Changes Associated with Transcriptional Noise
(A) Expression of cell-typical (INS for β-cells, GCG for α-cells) and non-typical hormone in cells, ranked by transcriptional noise. Dots represent individual cells, line is running mean, with k = n/5 (k = 69 for β cells and 199 for α cells). (B) Organismal age and expression of stress-related genes are strongly associated with transcriptional noise. All genes were tested for association with transcriptional noise (linear rank regression), shown are the top genes by coefficient, with FDR <1E-3. Heatmap shows loess fit. Rows marked with a black box indicate genes that are associated with response to stress (Yu et al., 2015; Daugaard et al., 2007; Paneni et al., 2013; Toone et al., 2001). See also Figure S2 and Table S4.
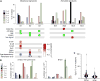
Figure 3. Somatic Mutation Profiles Derived from Single Primary Human Cells
(A) Substitution rates for each type of substitution in the three datasets. Somatic substitution rates were more than five times as high in pancreas as in brain (2.74 × 10−6 versus 0.52 × 10−6), whereas germline substitution rates were similar between the two. As expected, the rate of clonal substitutions in the tumor cell-line (GP5d) is several fold higher than germline rates in primary tissue. (B) Somatic substitutions are strongly enriched on untranslated regions compared to germline substitutions. Bars are mean ± SEM, n = 73. (C) Comparison of relative mutation rates of single-nucleotide substitutions in the context of the nucleotide immediately 5′ of the altered base. Different substitution types are separated by boxes with the substitution type indicated (e.g., C > A: C to A transversion). The relative substitution rate for C > T substitutions within a CpG context, and T > C substitutions is higher in brain than in the other tissues tested (p = 6.38E-61 and p = 1.89E-17, respectively; Wilcoxon test, n = 2,544 for pancreas, n = 73 for gp5d, and n = 332 for brain). (D) Detecting mRNA editing in brain samples. Shown is the number of splice site substitutions in the GRIA2 gene. T > C substitutions mapping to the transcribed (−) strand, corresponding to adenine substituted for guanine in the transcribed RNA, are highly enriched whereas other substitution types remain at baseline levels. Inlay shows mean number of GRIA2 substitutions per cell for the three datasets, brain is highly enriched in such substitutions (p = 5.40E-19. Bars are mean ± SEM, n = 2,544 for pancreas, n = 3323 for brain, and n = 73 for GP5d). See also Figure S3.
Figure 4. Mutational Signatures Derived from scRNA-Seq Data
(A) Single-nucleotide substitutions in 3,003 cells from pancreas, brain, and the colon cancer cell line GP5d were organized into mutational signatures using non-negative matrix factorization followed by agglomerative hierarchical clustering. Bar plot illustrates the percent of mutations attributed to each substitution type in each of the three signatures (S1–S3, left) and the four excluded signatures (SC1–SC4, right). Colors as in (A). Panel below the bar plot indicates selection items for determining whether to exclude the signature. Green, cause for inclusion; red, cause for exclusion. Bottom panel denotes the presence of a signature (columns) in a cell type (rows), with color scale indicating strength of signature as median substitution rate for cells of the indicated type. Blue boxes denote significant association between signature load and donor age. Bottom row indicates equivalent signatures from Alexandrov et al. (2013b). (B) Strand specificity differs between cell types. Mutations were annotated based on whether the mutated pyrimidine occurred on the transcribed (−) or untranscribed (+) strand. Bars represent mean ± SEM of raw substitution counts in endocrine cells (left) and brain cells (right). Note that endocrine cells have a strong strand bias for the transcribed strand for C > A, C > G, and C > T substitutions (p = 1.00E-79, 1.37e-28, and 6.40E-34, respectively; Wilcoxon test, n = 1,429) previously observed in oxidative stress-related tumor signatures, while brain has a bias for T > C substitutions on the transcribed strand (p = 3.41E-11; Wilcoxon test, n = 466) similar to tumor signature 12 (Alexandrov et al., 2013b). (C) Signature S2 is composed of two sub-signatures corresponding to cancer signatures 1 and 6. Violin plot show C > T substitutions with a preceding G as a fraction of all substitutions in a cell, which is a hallmark of cancer signature 6 and that separates GP5d and brain cells (p = 7.156E-11; Wilcoxon test, n = 73 for GP5d and n = 332 for brain cells). See also Figure S4 and Tables S6 and S7.
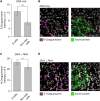
Figure 5. The Genomic DNA in Pancreatic Islets Are Highly Enriched in Oxidized Guanine
(A) Pancreatic β-cell DNA is enriched in oxidized guanosine. Nuclear staining intensity of anti 8-Oxoguanosine antibody was quantified for INS-positive or INS-negative cells, from the same images. Slides were treated with RNase so as to only measure oxidized bases on DNA. Bar plot indicates mean ± SEM (p = 7.30E-57; Wilcoxon test, n = 769 β-cells, 10,713 non-islet cells.). (B) Left: representative micrograph with 8-Oxoguanosine in magenta and nuclear stain (DAPI) in gray (scale bar, 50 µm). Right: insulin protein staining of the same region. Insulin-positive islet cell mass is at bottom left, boundary indicated with orange line. (C) Pancreatic β-cell RNA is marginally enriched in oxidized guanosine. Cytoplasmic staining intensity of anti 8-Oxoguanosine antibody was quantified for INS-positive β cells and INS-negative cells from the same slides. Bar plot indicates mean ± SEM (p = 9.5E-22, 1,239 β-cells, 21,048 surrounding cells). (D) Left: representative micrograph with 8-Oxoguanosine in magenta and nuclear stain (DAPI) in gray. Right: insulin protein staining of the same region. INS-positive islet cell mass boundary indicated with orange line. Scale bar, 50 µm. See also Figure S5.
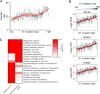
Figure 6. Transcriptional Correlates of Mutational Signatures
Endocrine pancreas cells were ordered according to the fraction of mutations attributed to Signature S1. (A) Average age is higher in cells with high S1 load (p = 5.95E-23, linear rank regression). Points are running mean, k = 10, and line is Loess fit, dotted lines indicate ± 0.999 confidence interval. (B) Each gene was tested for association with signature S1 (linear rank regression), shown are the top genes by coefficient, with p < 1E-15 (FDR corrected). Points are individual mRNA measurements, line loess fit as in (A). (C) Comparison of the top ten gene ontology (GO) categories positively correlated with signature S1 and transcriptional noise. Categories related to protein production, such as ribosomal proteins, recur in both. Color scale indicates FDR-adjusted p value, winsorized at 10−6. See also Table S5.
Comment in
- Transcriptional Noise and Somatic Mutations in the Aging Pancreas.
Swisa A, Kaestner KH, Dor Y. Swisa A, et al. Cell Metab. 2017 Dec 5;26(6):809-811. doi: 10.1016/j.cmet.2017.11.009. Cell Metab. 2017. PMID: 29211979
Similar articles
- Transcriptional Noise and Somatic Mutations in the Aging Pancreas.
Swisa A, Kaestner KH, Dor Y. Swisa A, et al. Cell Metab. 2017 Dec 5;26(6):809-811. doi: 10.1016/j.cmet.2017.11.009. Cell Metab. 2017. PMID: 29211979 - Aging and genome maintenance.
Vijg J, Busuttil RA, Bahar R, Dollé ME. Vijg J, et al. Ann N Y Acad Sci. 2005 Dec;1055:35-47. doi: 10.1196/annals.1323.007. Ann N Y Acad Sci. 2005. PMID: 16387716 Review. - CellBIC: bimodality-based top-down clustering of single-cell RNA sequencing data reveals hierarchical structure of the cell type.
Kim J, Stanescu DE, Won KJ. Kim J, et al. Nucleic Acids Res. 2018 Nov 30;46(21):e124. doi: 10.1093/nar/gky698. Nucleic Acids Res. 2018. PMID: 30102368 Free PMC article. - Transcriptome analysis of pancreatic cells across distant species highlights novel important regulator genes.
Tarifeño-Saldivia E, Lavergne A, Bernard A, Padamata K, Bergemann D, Voz ML, Manfroid I, Peers B. Tarifeño-Saldivia E, et al. BMC Biol. 2017 Mar 21;15(1):21. doi: 10.1186/s12915-017-0362-x. BMC Biol. 2017. PMID: 28327131 Free PMC article. - Impact of genome instability on transcription regulation of aging and senescence.
Vijg J. Vijg J. Mech Ageing Dev. 2004 Oct-Nov;125(10-11):747-53. doi: 10.1016/j.mad.2004.07.004. Mech Ageing Dev. 2004. PMID: 15541769 Review.
Cited by
- A cellular identity crisis? Plasticity changes during aging and rejuvenation.
Gorelov R, Hochedlinger K. Gorelov R, et al. Genes Dev. 2024 Oct 16;38(17-20):823-842. doi: 10.1101/gad.351728.124. Genes Dev. 2024. PMID: 39293862 Review. - The Human Islet: Mini-Organ With Mega-Impact.
Walker JT, Saunders DC, Brissova M, Powers AC. Walker JT, et al. Endocr Rev. 2021 Sep 28;42(5):605-657. doi: 10.1210/endrev/bnab010. Endocr Rev. 2021. PMID: 33844836 Free PMC article. Review. - Aggressive PDACs Show Hypomethylation of Repetitive Elements and the Execution of an Intrinsic IFN Program Linked to a Ductal Cell of Origin.
Espinet E, Gu Z, Imbusch CD, Giese NA, Büscher M, Safavi M, Weisenburger S, Klein C, Vogel V, Falcone M, Insua-Rodríguez J, Reitberger M, Thiel V, Kossi SO, Muckenhuber A, Sarai K, Lee AYL, Backx E, Zarei S, Gaida MM, Rodríguez-Paredes M, Donato E, Yen HY, Eils R, Schlesner M, Pfarr N, Hackert T, Plass C, Brors B, Steiger K, Weichenhan D, Arda HE, Rooman I, Kopp JL, Strobel O, Weichert W, Sprick MR, Trumpp A. Espinet E, et al. Cancer Discov. 2021 Mar;11(3):638-659. doi: 10.1158/2159-8290.CD-20-1202. Epub 2020 Oct 15. Cancer Discov. 2021. PMID: 33060108 Free PMC article. - Single-cell transcriptomics of human islet ontogeny defines the molecular basis of β-cell dedifferentiation in T2D.
Avrahami D, Wang YJ, Schug J, Feleke E, Gao L, Liu C; HPAP Consortium; Naji A, Glaser B, Kaestner KH. Avrahami D, et al. Mol Metab. 2020 Dec;42:101057. doi: 10.1016/j.molmet.2020.101057. Epub 2020 Jul 30. Mol Metab. 2020. PMID: 32739450 Free PMC article. - De novo identification of expressed cancer somatic mutations from single-cell RNA sequencing data.
Zhang T, Jia H, Song T, Lv L, Gulhan DC, Wang H, Guo W, Xi R, Guo H, Shen N. Zhang T, et al. Genome Med. 2023 Dec 18;15(1):115. doi: 10.1186/s13073-023-01269-1. Genome Med. 2023. PMID: 38111063 Free PMC article.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013b;500:415–421. - PMC - PubMed
MeSH terms
Grants and funding
- UC4 DK104211/DK/NIDDK NIH HHS/United States
- U01 HL099995/HL/NHLBI NIH HHS/United States
- U01 HL099999/HL/NHLBI NIH HHS/United States
- T32 DK007217/DK/NIDDK NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
- P30 DK116074/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials