A chemical and biological toolbox for Type Vd secretion: Characterization of the phospholipase A1 autotransporter FplA from Fusobacterium nucleatum - PubMed (original) (raw)
A chemical and biological toolbox for Type Vd secretion: Characterization of the phospholipase A1 autotransporter FplA from Fusobacterium nucleatum
Michael A Casasanta et al. J Biol Chem. 2017.
Abstract
Fusobacterium nucleatum is an oral pathogen that is linked to multiple human infections and colorectal cancer. Strikingly, F. nucleatum achieves virulence in the absence of large, multiprotein secretion systems (Types I, II, III, IV, and VI), which are widely used by Gram-negative bacteria for pathogenesis. By contrast, F. nucleatum strains contain genomic expansions of Type V secreted effectors (autotransporters) that are critical for host cell adherence, invasion, and biofilm formation. Here, we present the first characterization of an F. nucleatum Type Vd phospholipase class A1 autotransporter (strain ATCC 25586, gene FN1704) that we hereby rename Fusobacterium phospholipase autotransporter (FplA). Biochemical analysis of multiple Fusobacterium strains revealed that FplA is expressed as a full-length 85-kDa outer membrane-embedded protein or as a truncated phospholipase domain that remains associated with the outer membrane. Whereas the role of Type Vd secretion in bacteria remains unidentified, we show that FplA binds with high affinity to host phosphoinositide-signaling lipids, revealing a potential role for this enzyme in establishing an F. nucleatum intracellular niche. To further analyze the role of FplA, we developed an fplA gene knock-out strain, which will guide future in vivo studies to determine its potential role in F. nucleatum pathogenesis. In summary, using recombinant FplA constructs, we have identified a biochemical toolbox that includes lipid substrates for enzymatic assays, potent inhibitors, and chemical probes to detect, track, and characterize the role of Type Vd secreted phospholipases in Gram-negative bacteria.
Keywords: Fusobacterium; autotransporter; chemical biology; colorectal cancer; host-pathogen interaction; phospholipase A; protein secretion.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
Figure 1.
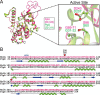
Fig. 1 FplA is a Type Vd autotransporter phospholipase from F. nucleatum. A, schematic representation of FplA domains and their location in the periplasm, outer membrane, and surface exposure of the PLA1 domain. Experimental data show FplA to be cleaved in a select set of F. nucleatum species, but the phospholipase domain remains associated with the bacterium. B, structure prediction of FplA domains: modeled patatin domain (residues 60–341) from PDB entry 5FYA (PlpD from P. aeruginosa); modeled β barrel (residues 432–759) and POTRA domain (residues 359–431) from PDB entry 4K3C (BamA from H. ducreyi).
Figure 2.
A predicted FplA structure is homologous to PlpD. A, alignment of a predicted FplA PLA1 domain structure with the crystal structure (PDB entry 5FQU) of the homologous phospholipase A1 enzyme PlpD from P. aeruginosa, with a magnified view of the catalytic dyad (Ser-98, Asp-243) and oxyanion hole (Gly-69, Gly-70, Gly-71). B, alignment of amino acids from PlpD (black) and FplA (pink) from the predicted structures of the PLA1 domains. The dashed red line indicates that this region was not predicted in the structure.
Figure 3.
Alignment of predicted FplA patatin domain structure with characterized phospholipase virulence factors. A, predicted FplA structure (residues 60–431) aligned with ExoU (P. aeruginosa) (PDB entry 4AKX) and VipD (L. pneumophila) (PDB entry 4AKF). B, zoomed in view of active sites after alignment showing similar architectures and residue placement of the catalytic dyad (Ser and Asp) and oxyanion hole (Gly, Gly, and Gly).
Figure 4.
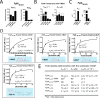
Characterization of FplA lipase activity with multiple fluorescent substrates. A, enzymatic assays show that FplA is a PLA1-specific enzyme with no PLA2 activity. B, steady-state kinetics of multiple FplA constructs with 4-MuH. C, FplA(20–431) _K_cat and Km values vary greatly across substrates with various acyl chain lengths. D and E, characterization of FplA enzyme kinetics.
Figure 5.
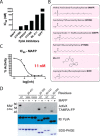
Characterization of FplA inhibitors. A, IC50 assays showing varying degrees of inhibition toward FplA by inhibitors previously shown to inhibit a variety of lipases. B, structure and names of tested inhibitors. C, IC50 plot of MAFP, the most potent (11 n
m
) FplA inhibitor characterized. D, analysis of the active site of FplA shows that the active site serine (Ser-98) reacts with ActivX TAMRA-FP probe but does not bind in the presence of the competitive inhibitor MAFP. S98A and D243A mutants will not bind the serine active site probe. Western blotting (IB) and SDS-polyacrylamide gels stained with Coomassie Blue serve as load controls for all constructs.
Figure 6.
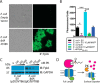
Expression of full-length FplA in E. coli and functional analysis. A, an OmpA(1–27) signal sequence allows for robust expression of FplA(20–760) on the surface of E. coli as seen by fluorescence microscopy with an anti-FplA antibody. B, enzymatic activity of FplA when live E. coli is added to reactions containing 4-MuH as a fluorescent substrate. C, PK, a cell-impenetrable nonspecific protease, is able to digest surface-exposed FplA, but not the cytoplasmic protein GAPDH. D, schematic of PK cleavage of full-length FplA from the surface of E. coli. EV, empty vector. Statistical analysis was performed using a multiple-comparison analysis by one-way analysis of variance. *, p ≤ 0.05; ***, p ≤ 0.0005. Error bars, S.D. IB, immunoblotting.
Figure 7.
Creation of an F. nucleatum 23726 Δ_fplA_. A, pDJSVT100 is a single-crossover integration plasmid for disruption of the fplA gene. Primers are labeled in red for PCRs A and B to confirm plasmid integration and gene knock-out. B, PCR confirmation of the F. nucleatum 23726 Δ_fplA_ strain. C, analysis of FplA protein (85.6 kDa) in WT and Δ_fplA_ by fluorescent chemical probe (ActivX TAMRA-FP) to label all active site serine enzymes in the bacteria (also serves as a load control), followed by transfer to PVDF for Western blot analysis by probing with an anti-FplA antibody.
Figure 8.
Western blot analysis of FplA in multiple Fusobacterium strains. A, initial characterization of FplA expression and protein size in mid-exponential phase (_A_600 = 0.7) and stationary phase (_A_600 = 1.2) shows that several strains produce a truncated form of FpA that consists of the PLA1 domain to which the FplA antibody was raised. B, Western blotting of medium from Fusobacterium growths shows that while truncated, FplA is not released into the medium and remains associated with the bacteria. C, analysis of FplA expression during early exponential phase growth (_A_600 = 0.2) reveals that strain 10953, which is cleaved in mid-exponential and stationary phase, is still in full-length state with a portion beginning to be cleaved. D, sequence alignment reveals that all FplA sequences from cleaved strains contain a highly charged motif at the PLA1/POTRA hinge region as a potential site for an unidentified protease, with the exception being the non-cleaved FplA proteins from 23726 and 25586, which contain a drastically different neutral motif. IB, immunoblotting.
Figure 9.
FplA binds with high affinity to phosphoinositide-signaling lipids that are critical for multiple cellular processes in a human host.
Similar articles
- Utilizing Whole Fusobacterium Genomes To Identify, Correct, and Characterize Potential Virulence Protein Families.
Umaña A, Sanders BE, Yoo CC, Casasanta MA, Udayasuryan B, Verbridge SS, Slade DJ. Umaña A, et al. J Bacteriol. 2019 Nov 5;201(23):e00273-19. doi: 10.1128/JB.00273-19. Print 2019 Dec 1. J Bacteriol. 2019. PMID: 31501282 Free PMC article. - Identification and characterization of fusolisin, the Fusobacterium nucleatum autotransporter serine protease.
Doron L, Coppenhagen-Glazer S, Ibrahim Y, Eini A, Naor R, Rosen G, Bachrach G. Doron L, et al. PLoS One. 2014 Oct 30;9(10):e111329. doi: 10.1371/journal.pone.0111329. eCollection 2014. PLoS One. 2014. PMID: 25357190 Free PMC article. - Quantitative Proteomic Analysis of Outer Membrane Vesicles from Fusobacterium nucleatum Cultivated in the Mimic Cancer Environment.
Zhang X, Wang Y, Fan R, Zhang L, Li Z, Zhang Y, Zheng W, Wang L, Liu B, Quan C. Zhang X, et al. Microbiol Spectr. 2023 Aug 17;11(4):e0039423. doi: 10.1128/spectrum.00394-23. Epub 2023 Jun 21. Microbiol Spectr. 2023. PMID: 37341631 Free PMC article. - Fusobacterium nucleatum: an emerging gut pathogen?
Allen-Vercoe E, Strauss J, Chadee K. Allen-Vercoe E, et al. Gut Microbes. 2011 Sep 1;2(5):294-8. doi: 10.4161/gmic.2.5.18603. Epub 2011 Sep 1. Gut Microbes. 2011. PMID: 22067936 Review. - Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer.
Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H, Maruyama R, Imai K, Yamamoto H, Shinomura Y. Nosho K, et al. World J Gastroenterol. 2016 Jan 14;22(2):557-66. doi: 10.3748/wjg.v22.i2.557. World J Gastroenterol. 2016. PMID: 26811607 Free PMC article. Review.
Cited by
- The patatin-like protein PlpD forms structurally dynamic homodimers in the Pseudomonas aeruginosa outer membrane.
Hanson SE, Dowdy T, Larion M, Doyle MT, Bernstein HD. Hanson SE, et al. Nat Commun. 2024 May 23;15(1):4389. doi: 10.1038/s41467-024-48756-6. Nat Commun. 2024. PMID: 38782915 Free PMC article. - A unified model for BAM function that takes into account type Vc secretion and species differences in BAM composition.
Leo JC, Linke D. Leo JC, et al. AIMS Microbiol. 2018 Jun 22;4(3):455-468. doi: 10.3934/microbiol.2018.3.455. eCollection 2018. AIMS Microbiol. 2018. PMID: 31294227 Free PMC article. - Type V Secretion in Gram-Negative Bacteria.
Bernstein HD. Bernstein HD. EcoSal Plus. 2019 Feb;8(2):10.1128/ecosalplus.ESP-0031-2018. doi: 10.1128/ecosalplus.ESP-0031-2018. EcoSal Plus. 2019. PMID: 30838971 Free PMC article. - RNA landscape of the emerging cancer-associated microbe Fusobacterium nucleatum.
Ponath F, Tawk C, Zhu Y, Barquist L, Faber F, Vogel J. Ponath F, et al. Nat Microbiol. 2021 Aug;6(8):1007-1020. doi: 10.1038/s41564-021-00927-7. Epub 2021 Jul 8. Nat Microbiol. 2021. PMID: 34239075 - Fusobacterium nucleatum - symbiont, opportunist and oncobacterium.
Brennan CA, Garrett WS. Brennan CA, et al. Nat Rev Microbiol. 2019 Mar;17(3):156-166. doi: 10.1038/s41579-018-0129-6. Nat Rev Microbiol. 2019. PMID: 30546113 Free PMC article. Review.
References
- Signat B., Roques C., Poulet P., and Duffaut D. (2011) Fusobacterium nucleatum in periodontal health and disease. Curr. Issues Mol. Biol. 13, 25–36 - PubMed
- Abed J., Emgård J. E., Zamir G., Faroja M., Almogy G., Grenov A., Sol A., Naor R., Pikarsky E., Atlan K. A., Mellul A., Chaushu S., Manson A. L., Earl A. M., Ou N., et al. (2016) Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 20, 215–225 - PMC - PubMed
- Dahya V., Patel J., Wheeler M., and Ketsela G. (2015) Fusobacterium nucleatum endocarditis presenting as liver and brain abscesses in an immunocompetent patient. Am. J. Med. Sci. 349, 284–285 - PubMed
- Rashidi A., Tahhan S. G., Cohee M. W., and Goodman B. M. (2012) Fusobacterium nucleatum infection mimicking metastatic cancer. Indian J. Gastroenterol. 31, 198–200 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials