Ecogenomics of virophages and their giant virus hosts assessed through time series metagenomics - PubMed (original) (raw)
Ecogenomics of virophages and their giant virus hosts assessed through time series metagenomics
Simon Roux et al. Nat Commun. 2017.
Abstract
Virophages are small viruses that co-infect eukaryotic cells alongside giant viruses (Mimiviridae) and hijack their machinery to replicate. While two types of virophages have been isolated, their genomic diversity and ecology remain largely unknown. Here we use time series metagenomics to identify and study the dynamics of 25 uncultivated virophage populations, 17 of which represented by complete or near-complete genomes, in two North American freshwater lakes. Taxonomic analysis suggests that these freshwater virophages represent at least three new candidate genera. Ecologically, virophage populations are repeatedly detected over years and evolutionary stable, yet their distinct abundance profiles and gene content suggest that virophage genera occupy different ecological niches. Co-occurrence analyses reveal 11 virophages strongly associated with uncultivated Mimiviridae, and three associated with eukaryotes among the Dinophyceae, Rhizaria, Alveolata, and Cryptophyceae groups. Together, these findings significantly augment virophage databases, help refine virophage taxonomy, and establish baseline ecological hypotheses and tools to study virophages in nature.Virophages are recently-identified small viruses that infect larger viruses, yet their diversity and ecological roles are poorly understood. Here, Roux and colleagues present time series metagenomics data revealing new virophage genera and their putative ecological interactions in two freshwater lakes.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Fig. 1
Phylogeny and summarized gene content of virophage genomes. The maximum-likelihood phylogenetic tree was computed from a concatenated alignment of four core genes (major and minor capsid proteins, DNA packaging enzyme, and Cysteine protease). SH-like support are indicated on nodes, and branches with < 50% support are displayed as multifurcations. The detection of key virophage genes is indicated for each genome on the right side. These key genes are classified as “core” (detected in all but one genome), ‘near-core’ (detected in > 68% of virophage genomes), and ‘signature genes’, i.e., genes specific to and detected in all members of known and newly proposed virophage genera. A question mark denotes the absence of genes similar to the DNA packaging enzyme in contig TBE_1002136, which is likely due to this genome being only a partial assembly, based on the comparison of this genome with the closely related TBE_1000887 (Supplementary Fig. 3). Six core genes previously identified from the comparison of isolate virophage genomes are highlighted with a star. DSLV, Dishui Lake virophage; OLV, Organic Lake virophage; QLV, Qinghai Lake virophage; SRHV, Sheep Rumen hybrid virophage; TBE, Trout Bog epilimnion; TBH, Trout Bog hypolimnion; YSLV, Yellowstone Lake virophage
Fig. 2
Virophage genome comparison and overall gene content. a Comparison of assembled genomes from the three new candidate virophage genera proposed. Tblastx similarities are depicted between genomes within each candidate genus. Circular contigs are indicated with a black dot next to the contig name. b Gene content of newly assembled freshwater virophages. Predicted genes were first affiliated to previously published virophages, then to other viral and cellular genomes, and finally to other virophages assembled in Lake Mendota and Trout Bog Lake. ORFs with no significant similarity to any database were considered ORFan. DSLV, Dishui Lake virophage; YSLV, Yellowstone Lake virophage
Fig. 3
SNPs density of virophage genomes (a) and location of non-syonymous SNPs on Mendota_1002202 minor capsid protein (b, c). a Virophages are colored according to the lake where they were identified. The two virophage genomes encoding divergent family B DNA polymerase genes are highlighted with a bold outline. b Subset of the sputnik virion model (3J26), with major and minor capsid proteins depicted in gray and blue, respectively. The model is oriented so that the bottom of the picture correspond to the inside of the virion, and the top to the outside. c The structure of a single minor capsid protein unit (from the same sputnik model 3J26), is colored by N to C orientation (from blue N-terminal to red C-terminal). The location of non-synonymous SNPs observed in Mendota_1002202 are indicated with gray spheres
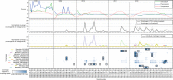
Fig. 4
Virophage abundance and the environmental data in Lake Mendota. Pigment concentrations and temperature are presented as Z-scores to focus on their variations across the time series rather than their absolute values. Virophage abundance are based on the mapping of metagenomic reads to virophage contigs. Individual virophage coverage is plotted in the third panel and highlighted with a black outline in the heatmap (fourth panel) when >75% of the contig is covered
Fig. 5
Virophage abundance, chlorophyll a concentration, tempreature, and dissolved organic carbon in Trout Bog Lake Epi- and Hypolimnion. Chlorophyll a, temperature, and DOC levels are displayed as Z-score to highlight their variation across each data set. Temperature for Trout Bog Lake Hypolimnion was stable at 4.6 °C (standard deviation: ± 0.7 °C). Virophage abundance are based on the mapping of metagenomic reads to virophage contigs. Individual virophage coverage is plotted in the third panel and virophages are highlighted with a black outline in the heatmap (fourth panel) when > 75% of the contig is covered
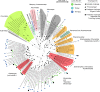
Fig. 6
NCLDV diversity in Lake Mendota and Trout Bog Lake, and predicted associations to virophages. The maximum-likelihood tree was computed from a DNA PolB multiple alignment. All branches with support <0.5 were collapsed. NCLDV clades are highlighted in colors, and associated with the host range based on the clade isolated members. Metagenomic sequences from Lake Mendota and Trout Bog Lake are highlighted with a colored circle. NCLDV associated with virophages are indicated with black shapes (either isolated virophages, the ‘provirophage’ from Phaeocystis globosa virus 16T, Organic Lake NCLDV, or metagenomic sequences from Lake Mendota and Trout Bog Lake co-occurring with virophage contigs)
Similar articles
- Novel Cell-Virus-Virophage Tripartite Infection Systems Discovered in the Freshwater Lake Dishui Lake in Shanghai, China.
Xu S, Zhou L, Liang X, Zhou Y, Chen H, Yan S, Wang Y. Xu S, et al. J Virol. 2020 May 18;94(11):e00149-20. doi: 10.1128/JVI.00149-20. Print 2020 May 18. J Virol. 2020. PMID: 32188734 Free PMC article. - Three novel virophage genomes discovered from Yellowstone Lake metagenomes.
Zhou J, Sun D, Childers A, McDermott TR, Wang Y, Liles MR. Zhou J, et al. J Virol. 2015 Jan 15;89(2):1278-85. doi: 10.1128/JVI.03039-14. Epub 2014 Nov 12. J Virol. 2015. PMID: 25392206 Free PMC article. - The Virophage Family Lavidaviridae.
Fischer MG. Fischer MG. Curr Issues Mol Biol. 2021;40:1-24. doi: 10.21775/cimb.040.001. Epub 2020 Feb 24. Curr Issues Mol Biol. 2021. PMID: 32089519 Review. - The Expanding Family of Virophages.
Bekliz M, Colson P, La Scola B. Bekliz M, et al. Viruses. 2016 Nov 23;8(11):317. doi: 10.3390/v8110317. Viruses. 2016. PMID: 27886075 Free PMC article. Review.
Cited by
- Astrovirology: how viruses enhance our understanding of life in the Universe.
Trubl G, Stedman KM, Bywaters KF, Matula EE, Sommers P, Roux S, Merino N, Yin J, Kaelber JT, Avila-Herrera A, Johnson PA, Johnson JC, Borges S, Weber PK, Pett-Ridge J, Boston PJ. Trubl G, et al. Int J Astrobiol. 2023 Aug;22(4):247-271. doi: 10.1017/s1473550423000058. Epub 2023 Apr 5. Int J Astrobiol. 2023. PMID: 38046673 Free PMC article. - Coupled laboratory and field investigations resolve microbial interactions that underpin persistence in hydraulically fractured shales.
Borton MA, Hoyt DW, Roux S, Daly RA, Welch SA, Nicora CD, Purvine S, Eder EK, Hanson AJ, Sheets JM, Morgan DM, Wolfe RA, Sharma S, Carr TR, Cole DR, Mouser PJ, Lipton MS, Wilkins MJ, Wrighton KC. Borton MA, et al. Proc Natl Acad Sci U S A. 2018 Jul 10;115(28):E6585-E6594. doi: 10.1073/pnas.1800155115. Epub 2018 Jun 25. Proc Natl Acad Sci U S A. 2018. PMID: 29941576 Free PMC article. - Metaviromics coupled with phage-host identification to open the viral 'black box'.
Moon K, Cho JC. Moon K, et al. J Microbiol. 2021 Mar;59(3):311-323. doi: 10.1007/s12275-021-1016-9. Epub 2021 Feb 23. J Microbiol. 2021. PMID: 33624268 Review. - Metagenomic Analysis of the Diversity of DNA Viruses in the Surface and Deep Sea of the South China Sea.
Liang Y, Wang L, Wang Z, Zhao J, Yang Q, Wang M, Yang K, Zhang L, Jiao N, Zhang Y. Liang Y, et al. Front Microbiol. 2019 Aug 23;10:1951. doi: 10.3389/fmicb.2019.01951. eCollection 2019. Front Microbiol. 2019. PMID: 31507563 Free PMC article. - Freshwater carbon and nutrient cycles revealed through reconstructed population genomes.
Linz AM, He S, Stevens SLR, Anantharaman K, Rohwer RR, Malmstrom RR, Bertilsson S, McMahon KD. Linz AM, et al. PeerJ. 2018 Dec 10;6:e6075. doi: 10.7717/peerj.6075. eCollection 2018. PeerJ. 2018. PMID: 30581671 Free PMC article.