Impaired HLA Class I Antigen Processing and Presentation as a Mechanism of Acquired Resistance to Immune Checkpoint Inhibitors in Lung Cancer - PubMed (original) (raw)
. 2017 Dec;7(12):1420-1435.
doi: 10.1158/2159-8290.CD-17-0593. Epub 2017 Oct 12.
Jungmin Choi 3, Katherine Hastings 2, Anna Truini 2, Ila Datar 4, Ryan Sowell 5, Anna Wurtz 2, Weilai Dong 3, Guoping Cai 4, Mary Ann Melnick 2, Victor Y Du 5, Joseph Schlessinger 2 6, Sarah B Goldberg 7 2, Anne Chiang 7 2, Miguel F Sanmamed 5, Ignacio Melero 8 9, Jackeline Agorreta 8 9, Luis M Montuenga 8 9, Richard Lifton 3, Soldano Ferrone 10, Paula Kavathas 2 5 11, David L Rimm 2 4, Susan M Kaech 2 5, Kurt Schalper 7 2 4, Roy S Herbst 7 2 6, Katerina Politi 1 2 4
Affiliations
- PMID: 29025772
- PMCID: PMC5718941
- DOI: 10.1158/2159-8290.CD-17-0593
Impaired HLA Class I Antigen Processing and Presentation as a Mechanism of Acquired Resistance to Immune Checkpoint Inhibitors in Lung Cancer
Scott Gettinger et al. Cancer Discov. 2017 Dec.
Abstract
Mechanisms of acquired resistance to immune checkpoint inhibitors (ICI) are poorly understood. We leveraged a collection of 14 ICI-resistant lung cancer samples to investigate whether alterations in genes encoding HLA Class I antigen processing and presentation machinery (APM) components or interferon signaling play a role in acquired resistance to PD-1 or PD-L1 antagonistic antibodies. Recurrent mutations or copy-number changes were not detected in our cohort. In one case, we found acquired homozygous loss of B2M that caused lack of cell-surface HLA Class I expression in the tumor and a matched patient-derived xenograft (PDX). Downregulation of B2M was also found in two additional PDXs established from ICI-resistant tumors. CRISPR-mediated knockout of B2m in an immunocompetent lung cancer mouse model conferred resistance to PD-1 blockade in vivo, proving its role in resistance to ICIs. These results indicate that HLA Class I APM disruption can mediate escape from ICIs in lung cancer.Significance: As programmed death 1 axis inhibitors are becoming more established in standard treatment algorithms for diverse malignancies, acquired resistance to these therapies is increasingly being encountered. Here, we found that defective antigen processing and presentation can serve as a mechanism of such resistance in lung cancer. Cancer Discov; 7(12); 1420-35. ©2017 AACR. This article is highlighted in the In This Issue feature, p. 1355.
©2017 American Association for Cancer Research.
Conflict of interest statement
The authors have the following conflicts to disclose:
- Research funding from AstraZeneca (SBG, KP, SMK, DLR), Roche (KP, SMK, IM), Kolltan (KP), BMS (IM), Pfizer (IM), Cepheid (DLR), Navigate/Novartis (DLR), Gilead Sciences (DLR), Pierre Fabre (DLR), Perkin Elmer (DLR).
- Consulting/Advisory Role honoraria from AstraZeneca (SBG, AC, KP, IM, DLR), Roche (IM), Merck-Serono (IM), Lilly (IM), BMS (IM, DLR), Agendia (DLR), Bethyl Labs (DLR), Biocept (DLR), Cell Signaling Technology (DLR), Merck (KP, DLR), Novartis (KP), OptraScan (DLR), Perkin Elmer (DLR), Ultivue (DLR).
- Equity in Metamark Genetics (DLR) and PixelGear (DLR).
- Royalties in IP licensed from MSKCC to Molecular MD (KP).
Figures
Figure 1. Analytical process and characteristics of the cohort of cases of acquired resistance to immune checkpoint inhibitors
(A) Schematic representation of the repeat biopsy program and sample analysis. Tumor specimens (and corresponding PDXs when available) collected at the time of resistance to ICIs and before treatment with immune checkpoint inhibitors along with germline DNA were analyzed using whole exome sequencing. For select samples with sufficient material, RNA sequencing and quantitative immunofluorescence were also performed. (B) Pie-chart illustrating the types of therapies received by patients in this study. (C) Swimmer’s plot indicating time of response, resistance to ICIs, and length of time on therapy for individual patients.
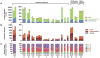
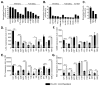
Figure 2. Mutational landscape of acquired resistance to immune checkpoint inhibitors
Bar graphs showing the (A) somatic mutation burden (B) neoantigen burden and (C) mutational signatures in pre-immunotherapy specimens (P, P1), immunotherapy resistant specimens (IR) and xenografts (X).
Figure 3. The genomic landscape of antigen processing and presentation pathway genes
Oncoprint generated from whole exome sequencing data from 13 cases. Only genes with alterations are shown. Mutated genes are listed vertically in order of frequency of somatic single nucleotide mutations or copy number alterations in the pre-immunotherapy cases. A case with a hypermutator phenotype (#17) sample was excluded from this analysis. Pre-immunotherapy specimens are shown on the left and immunotherapy resistant tumors on the right. *Of note, sample 26IR1 was collected from a site (adrenal metastasis) that responded to a short course of ICI, but progressed during a 2-month delay of treatment when steroids were administered for cerebral edema associated with new intracranial disease (sample 26IR3) and pneumonitis. This site subsequently regressed with re-introduction of ICI.
Figure 4. B2M loss in a case of acquired resistance to immunotherapy
(A) Treatment timeline for case #23. After receiving palliative thoracic irradiation and 2 lines of standard chemotherapy, the patient initiated trial therapy with anti-PD-L1 andanti-CTLA-4 agents. First on-trial imaging assessment demonstrated partial response. Imaging after 4 months of therapy showed a new celiac mass with sustained response at known sites of disease. After resection of this mass, this patient continued trial therapy, completing the prescribed one-year course, during which the patient had no further progression of disease. (B, C) Hematoxylin and eosin, CD8 and PD-L1 immunohistochemical staining of the tumors before treatment and at acquired resistance to anti-PD-L1 and anti-CTLA-4 blockade. (D) Copy number variation (CNV) analysis of paired tumor specimens and a PDX derived from the immunotherapy resistant tumor specimen revealed acquired homozygous loss of B2M at the time of acquired resistance to anti-PD-L1 and anti-CTLA-4 blockade. (E, F) Multiplex quantitative immunofluorescence (QIF) of B2M in pre-immunotherapy and immunotherapy resistant tumor tissues from case #23**. (E)** Bar graph depicting the levels of B2M in whole tumor sections from pre-immunotherapy and immunotherapy resistant samples measured using multiplex quantitative immunofluorescence and quantified with AQUA® software. Numbers in the individual bars represent total fields of view analyzed. Statistical significance was calculated using the Mann-Whitney test. ****p<0.0001. AU, arbitrary units. (F) Multiplex immunofluorescence image representing one field-of-view (FOV), showing the expression of B2M (purple) specifically in the tumor compartment represented by cytokeratin positive epithelial cells (green) and nuclear staining with DAPI (blue) respectively.
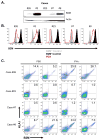
Figure 5. Defects in HLA class I antigen processing and presentation in ICI-resistant tumors following B2M loss
(A) Western blot analysis showing the absence of B2M protein expression in a patient derived xenograft (PDX) from case #23 generated at the time of acquired resistance to anti-PD-L1 and anti-CTLA-4 blockade. Also shown is B2M protein expression in 4 PDXs established from other patients with tumors resistant to PD-1 pathway blockade (cases #26, 3, 7, and 8). (B) Flow cytometry analysis for expression of B2M on PDX samples from cases #26, 3, 23, 7, and 8. (C) Flow cytometry analysis of PDXs derived from cases #23, #26, #8 and #7 for B2M and HLA-I cell surface expression following intratumoral injection of either PBS or IFNγ. Each flow plot represents an independent mouse tumor used for these studies.
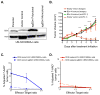
Figure 6. B2m loss confers resistance to anti-PD-1 therapy and impairs CD8 cytotoxicity in vivo
(A) Western blot analysis of B2m expression in UN-SCC680AJ lung cancer cells transfected with a plasmid expressing Cas9 and a sgRNA targeting B2m (sgB2m polyclonal). Additionally, single cell sorting identified clonal populations where B2m expression was absent (sgB2m monoclonal). (B) 5×105 UN-SCC680AJ cells from the monoclonal B2m null population and vector control cells were injected into the dorsal flanks of A/J mice subcutaneously. Tumors were allowed to grow to approximately 30 mm3 before administration of 100 μg of anti-PD-1 or isotype control via intraperitoneal injection every 3 days. Tumors were measured every 3 days with a caliper, and tumor volumes were calculated. Data are presented as the mean tumor volume ± SEM. The size of the tumors in the B2m wild-type and B2m knock-out lines treated with anti-PD1 were compared at the indicated time-points using a T-test. ***p<0.001, ****p<0.0001. Additionally, cytotoxicity assays were performed to assess the percentage of viable target cells (Empty vector (EV) UNSCC680AJ or sgB2m UNSCC680AJ) after co-culturing the cell lines in vitro with EV UNSCC680AJ tumor-infiltrating CD8 T cells (C) or T cells from the spleen (D). Data represent n=2 mice, with error bars denoting standard error of the mean.
Figure 7. RNA sequencing and tumor immunoprofiling of T-cell inhibitory and activation markers in tumor tissues from patients pre-immunotherapy and at resistance to ICIs
Ratio of normalized RNA sequencing-derived read counts of T cell inhibitory and activating receptors (A), ligands (B), and effector molecules (C) in immunotherapy resistant specimens (n=7) compared to pre-immunotherapy specimens (n=4). (D–F) Quantitative immunofluorescence analysis showing the levels of the immune inhibitory molecule LAG-3 (D), the T cell exhaustion marker PD-1 (E), T-cell activation effector molecule Granzyme B (GZB) (F) and T-cell proliferation marker Ki-67 (G) in CD3 positive tumor infiltrating lymphocytes (TILs) in paired pre- and post-immunotherapy tumor samples as quantified using the AQUA® software. Numbers in the individual bars represent total fields of view analyzed. Statistical significance was calculated using the Mann-Whitney test. P1- pre-immunotherapy and IR- immunotherapy resistant. ****p<0.0001, ***p<0.001 **p<0.01, *p<0.05.
Similar articles
- Genomic Profiling of Patient-Derived Xenografts for Lung Cancer Identifies B2M Inactivation Impairing Immunorecognition.
Pereira C, Gimenez-Xavier P, Pros E, Pajares MJ, Moro M, Gomez A, Navarro A, Condom E, Moran S, Gomez-Lopez G, Graña O, Rubio-Camarillo M, Martinez-Martí A, Yokota J, Carretero J, Galbis JM, Nadal E, Pisano D, Sozzi G, Felip E, Montuenga LM, Roz L, Villanueva A, Sanchez-Cespedes M. Pereira C, et al. Clin Cancer Res. 2017 Jun 15;23(12):3203-3213. doi: 10.1158/1078-0432.CCR-16-1946. Epub 2017 Mar 16. Clin Cancer Res. 2017. PMID: 28302866 - Human Leukocyte Antigen Class I Antigen-Processing Machinery Upregulation by Anticancer Therapies in the Era of Checkpoint Inhibitors: A Review.
Sadagopan A, Michelakos T, Boyiadzis G, Ferrone C, Ferrone S. Sadagopan A, et al. JAMA Oncol. 2022 Mar 1;8(3):462-473. doi: 10.1001/jamaoncol.2021.5970. JAMA Oncol. 2022. PMID: 34940799 Free PMC article. Review. - The role of classical and non-classical HLA class I antigens in human tumors.
Bukur J, Jasinski S, Seliger B. Bukur J, et al. Semin Cancer Biol. 2012 Aug;22(4):350-8. doi: 10.1016/j.semcancer.2012.03.003. Epub 2012 Mar 24. Semin Cancer Biol. 2012. PMID: 22465194 Review. - Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma.
Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, Saco J, Homet Moreno B, Mezzadra R, Chmielowski B, Ruchalski K, Shintaku IP, Sanchez PJ, Puig-Saus C, Cherry G, Seja E, Kong X, Pang J, Berent-Maoz B, Comin-Anduix B, Graeber TG, Tumeh PC, Schumacher TN, Lo RS, Ribas A. Zaretsky JM, et al. N Engl J Med. 2016 Sep 1;375(9):819-29. doi: 10.1056/NEJMoa1604958. Epub 2016 Jul 13. N Engl J Med. 2016. PMID: 27433843 Free PMC article. - Immunological and clinical significance of HLA class I antigen processing machinery component defects in malignant cells.
Concha-Benavente F, Srivastava R, Ferrone S, Ferris RL. Concha-Benavente F, et al. Oral Oncol. 2016 Jul;58:52-8. doi: 10.1016/j.oraloncology.2016.05.008. Epub 2016 Jun 2. Oral Oncol. 2016. PMID: 27264839 Free PMC article. Review.
Cited by
- Prognostic Indicators for Precision Treatment of Non-Small Cell Lung Carcinoma.
Ghosh DD, McDonald H, Dutta R, Krishnan K, Thilakan J, Paul MK, Arya N, Rao M, Rangnekar VM. Ghosh DD, et al. Cells. 2024 Oct 28;13(21):1785. doi: 10.3390/cells13211785. Cells. 2024. PMID: 39513892 Free PMC article. Review. - Predictive biomarkers for immune checkpoint inhibitors therapy in lung cancer.
Yao J, Lin X, Zhang X, Xie M, Ma X, Bao X, Song J, Liang Y, Wang Q, Xue X. Yao J, et al. Hum Vaccin Immunother. 2024 Dec 31;20(1):2406063. doi: 10.1080/21645515.2024.2406063. Epub 2024 Oct 16. Hum Vaccin Immunother. 2024. PMID: 39415535 Free PMC article. Review. - Aberrant cytoplasmic expression of UHRF1 restrains the MHC-I-mediated anti-tumor immune response.
Tan L, Yin T, Xiang H, Wang L, Mudgal P, Chen J, Ding Y, Wang G, Lim BJW, Huang Y, Huang D, Liang Y, Alexander PB, Xiang K, Wang E, Yan C, Ma Z, Tan M, Li QJ, Wang XF. Tan L, et al. Nat Commun. 2024 Oct 3;15(1):8569. doi: 10.1038/s41467-024-52902-5. Nat Commun. 2024. PMID: 39362877 Free PMC article. - Immunoediting Dynamics in Glioblastoma: Implications for Immunotherapy Approaches.
Amin T, Hossain A, Jerin N, Mahmud I, Rahman MA, Rafiqul Islam SM, Islam SMBU. Amin T, et al. Cancer Control. 2024 Jan-Dec;31:10732748241290067. doi: 10.1177/10732748241290067. Cancer Control. 2024. PMID: 39353594 Free PMC article. Review. - Personalized prediction of immunotherapy response in lung cancer patients using advanced radiomics and deep learning.
Liao CY, Chen YM, Wu YT, Chao HS, Chiu HY, Wang TW, Chen JR, Shiao TH, Lu CF. Liao CY, et al. Cancer Imaging. 2024 Sep 30;24(1):129. doi: 10.1186/s40644-024-00779-4. Cancer Imaging. 2024. PMID: 39350284 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- R01 CA195720/CA/NCI NIH HHS/United States
- F32 CA210516/CA/NCI NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- P30 CA016359/CA/NCI NIH HHS/United States
- T32 HL007974/HL/NHLBI NIH HHS/United States
- P50 CA196530/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous