A Compendium of Co-regulated Protein Complexes in Breast Cancer Reveals Collateral Loss Events - PubMed (original) (raw)
A Compendium of Co-regulated Protein Complexes in Breast Cancer Reveals Collateral Loss Events
Colm J Ryan et al. Cell Syst. 2017.
Abstract
Protein complexes are responsible for the bulk of activities within the cell, but how their behavior and abundance varies across tumors remains poorly understood. By combining proteomic profiles of breast tumors with a large-scale protein-protein interaction network, we have identified a set of 285 high-confidence protein complexes whose subunits have highly correlated protein abundance across tumor samples. We used this set to identify complexes that are reproducibly under- or overexpressed in specific breast cancer subtypes. We found that mutation or deletion of one subunit of a co-regulated complex was often associated with a collateral reduction in protein expression of additional complex members. This collateral loss phenomenon was typically evident from proteomic, but not transcriptomic, profiles, suggesting post-transcriptional control. Mutation of the tumor suppressor E-cadherin (CDH1) was associated with a collateral loss of members of the adherens junction complex, an effect we validated using an engineered model of E-cadherin loss.
Keywords: E-cadherin; breast cancer; cancer; driver gene mutations; post-transcriptional control; protein complexes; proteogenomics; proteomics.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
BrCa-Core Complex Discovery (A) An integrated protein-protein interaction network is combined with tumor proteomic profiles to identify sets of densely connected proteins that display correlated expression profiles across tumor proteomes. By comparing the results with those derived from randomly relabeled protein interaction networks, we can estimate the false discovery rate (FDR). The BrCa-Core set contains 285 complexes at an estimated FDR of 10%. (B) BrCa-Core 17: the COP9 signalosome. The heatmap on the right shows protein expression of all subunits across 77 breast tumor proteomes. These have been sorted based on the mean abundance of all subunits. (C) BrCa-Core 14: the conserved oligomeric Golgi (COG) complex. (D) BrCa-Core 47 contains the ORC2–5 complex found in CORUM with the addition of LRWD1. (E) BrCa-Core 25, the COPI complex. (F) BrCa-Core 48, the EARP complex with the recently identified EARP interactor TSSC1. (G) BrCa-Core 26, the Commander complex.
Figure 2
Subtype-Specific Complex Expression (A) Heatmap displaying protein expression levels of specific BrCa-Core complexes. Tumor samples are grouped according to subtype (using IHC markers), indicated on top of the heatmap. Genes are grouped into specific complexes indicated on the right of the heatmap. Shown are the expression levels taken from Tyanova et al., 2016 (used for validation). These expression levels have been normalized such that the maximum expression level is 1 and minimum is 0. Heatmap for the discovery dataset (Mertins et al., 2016) is shown in Figure S2A. Complexes differentially expressed in specific subtypes are highlighted with boxes colored to match the subtype they are differentially expressed in. (B) Boxplots displaying the subtype-specific protein expression levels of selected subunits of the COPI complex (BrCa-Core 25) in the Tyanova et al. dataset (top) and TCGA dataset (bottom). These boxplots show median and interquartile range and are colored according to sample subtype (matching Figure 2A).
Figure 3
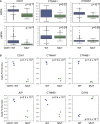
Subunit Loss Is Associated with a Reduction in Protein Complex Expression (A) Model displaying a potential series of events; mutation or deletion of one subunit is associated with reduced protein abundance of that subunit and potentially a reduction in expression of the entire complex. (B) Mutation or deletion of EXOC2 is associated with a reduction in protein abundance of the exocyst complex (BrCa-Core 27). Boxplots display the protein abundance of different subunits partitioned according to EXOC2 status. Each boxplot shows the median and interquartile range. Genes marked with a star indicate those whose proteomic abundance is significantly lower (one-sided Mann-Whitney test, p < 0.05) in samples with EXOC2 mutation/deletion. (C) CDH1 mutation is associated with a reduction in protein expression of an adherens junction complex (BrCa-Core 30). Legend as for (B).
Figure 4
E-cadherin Loss Is Associated with Reduced Expression of an Adherens Junction Complex (A) In tumor samples, CDH1 mutation is associated with a decrease in mRNA and protein expression of CDH1 but only of protein expression for CTNNA1 and CTNNB1. All expression and RPPA measurements are Z scores. Boxplots show median and interquartile range. p values calculated using a Mann-Whitney test. mRNA measurements for all three genes were available for 992 tumors, RPPA data for CDH1, and CTNNB1 were available for 760 tumors, while RPPA data for CTNNA1 were available for only 64 tumors. (B) Protein expression measured in a pair of isogenic MCF7 cell lines that differ by CDH1 status. Shown are the log2 label-free quantification intensities. p values are calculated using a two-sided heteroscedastic t test.
Comment in
- All for One, and One for All.
Kim E, Hart T. Kim E, et al. Cell Syst. 2017 Oct 25;5(4):314-316. doi: 10.1016/j.cels.2017.10.005. Cell Syst. 2017. PMID: 29073370
Similar articles
- Proteomics analysis of E-cadherin knockdown in epithelial breast cancer cells.
Vergara D, Simeone P, Latorre D, Cascione F, Leporatti S, Trerotola M, Giudetti AM, Capobianco L, Lunetti P, Rizzello A, Rinaldi R, Alberti S, Maffia M. Vergara D, et al. J Biotechnol. 2015 May 20;202:3-11. doi: 10.1016/j.jbiotec.2014.10.034. Epub 2014 Nov 4. J Biotechnol. 2015. PMID: 25449012 - The Cdc42/Rac1 regulator CdGAP is a novel E-cadherin transcriptional co-repressor with Zeb2 in breast cancer.
He Y, Northey JJ, Pelletier A, Kos Z, Meunier L, Haibe-Kains B, Mes-Masson AM, Côté JF, Siegel PM, Lamarche-Vane N. He Y, et al. Oncogene. 2017 Jun 15;36(24):3490-3503. doi: 10.1038/onc.2016.492. Epub 2017 Jan 30. Oncogene. 2017. PMID: 28135249 Free PMC article. - Widespread Post-transcriptional Attenuation of Genomic Copy-Number Variation in Cancer.
Gonçalves E, Fragoulis A, Garcia-Alonso L, Cramer T, Saez-Rodriguez J, Beltrao P. Gonçalves E, et al. Cell Syst. 2017 Oct 25;5(4):386-398.e4. doi: 10.1016/j.cels.2017.08.013. Epub 2017 Oct 11. Cell Syst. 2017. PMID: 29032074 Free PMC article. - Mutations of the human E-cadherin (CDH1) gene.
Berx G, Becker KF, Höfler H, van Roy F. Berx G, et al. Hum Mutat. 1998;12(4):226-37. doi: 10.1002/(SICI)1098-1004(1998)12:4<226::AID-HUMU2>3.0.CO;2-D. Hum Mutat. 1998. PMID: 9744472 Review. - The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression.
Berx G, Van Roy F. Berx G, et al. Breast Cancer Res. 2001;3(5):289-93. doi: 10.1186/bcr309. Epub 2001 Jun 28. Breast Cancer Res. 2001. PMID: 11597316 Free PMC article. Review.
Cited by
- Development of synthetic lethality in cancer: molecular and cellular classification.
Li S, Topatana W, Juengpanich S, Cao J, Hu J, Zhang B, Ma D, Cai X, Chen M. Li S, et al. Signal Transduct Target Ther. 2020 Oct 19;5(1):241. doi: 10.1038/s41392-020-00358-6. Signal Transduct Target Ther. 2020. PMID: 33077733 Free PMC article. Review. - A computational framework for the inference of protein complex remodeling from whole-proteome measurements.
Buljan M, Banaei-Esfahani A, Blattmann P, Meier-Abt F, Shao W, Vitek O, Tang H, Aebersold R. Buljan M, et al. Nat Methods. 2023 Oct;20(10):1523-1529. doi: 10.1038/s41592-023-02011-w. Epub 2023 Sep 25. Nat Methods. 2023. PMID: 37749212 Free PMC article. - MOLI: multi-omics late integration with deep neural networks for drug response prediction.
Sharifi-Noghabi H, Zolotareva O, Collins CC, Ester M. Sharifi-Noghabi H, et al. Bioinformatics. 2019 Jul 15;35(14):i501-i509. doi: 10.1093/bioinformatics/btz318. Bioinformatics. 2019. PMID: 31510700 Free PMC article. - CoExpresso: assess the quantitative behavior of protein complexes in human cells.
Chalabi MH, Tsiamis V, Käll L, Vandin F, Schwämmle V. Chalabi MH, et al. BMC Bioinformatics. 2019 Jan 9;20(1):17. doi: 10.1186/s12859-018-2573-8. BMC Bioinformatics. 2019. PMID: 30626316 Free PMC article. - E-Cadherin/ROS1 Inhibitor Synthetic Lethality in Breast Cancer.
Bajrami I, Marlow R, van de Ven M, Brough R, Pemberton HN, Frankum J, Song F, Rafiq R, Konde A, Krastev DB, Menon M, Campbell J, Gulati A, Kumar R, Pettitt SJ, Gurden MD, Cardenosa ML, Chong I, Gazinska P, Wallberg F, Sawyer EJ, Martin LA, Dowsett M, Linardopoulos S, Natrajan R, Ryan CJ, Derksen PWB, Jonkers J, Tutt ANJ, Ashworth A, Lord CJ. Bajrami I, et al. Cancer Discov. 2018 Apr;8(4):498-515. doi: 10.1158/2159-8290.CD-17-0603. Cancer Discov. 2018. PMID: 29610289 Free PMC article.
References
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. - PubMed
- Bader J.S., Chaudhuri A., Rothberg J.M., Chant J. Gaining confidence in high-throughput protein interaction networks. Nat. Biotechnol. 2004;22:78–85. - PubMed
- Benjamini Y., Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. Roy Stat. Soc. B Met. 1995;57:289–300.
Publication types
MeSH terms
Substances
Grants and funding
- 103049/Z/13/Z/WT_/Wellcome Trust/United Kingdom
- C347/A8363/CRUK_/Cancer Research UK/United Kingdom
- 14276/CRUK_/Cancer Research UK/United Kingdom
- 24439/CRUK_/Cancer Research UK/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous