Virus-Mediated Genome Editing via Homology-Directed Repair in Mitotic and Postmitotic Cells in Mammalian Brain - PubMed (original) (raw)
Virus-Mediated Genome Editing via Homology-Directed Repair in Mitotic and Postmitotic Cells in Mammalian Brain
Jun Nishiyama et al. Neuron. 2017.
Abstract
Precise genome editing via homology-directed repair (HDR) in targeted cells, particularly in vivo, provides an invaluable tool for biomedical research. However, HDR has been considered to be largely restricted to dividing cells, making it challenging to apply the technique in postmitotic neurons. Here we show that precise genome editing via HDR is possible in mature postmitotic neurons as well as mitotic cells in mice brain by combining CRISPR-Cas9-mediated DNA cleavage and the efficient delivery of donor template with adeno-associated virus (AAV). Using this strategy, we achieved efficient tagging of endogenous proteins in primary and organotypic cultures in vitro and developing, adult, aged, and pathological brains in vivo. Thus, AAV- and CRISPR-Cas9-mediated HDR will be broadly useful for precise genome editing in basic and translational neuroscience.
Keywords: AAV; CRISPR; Cas9; Genome editing; HDR; SLENDR; in vivo; neuron; postmitotic; vSLENDR.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
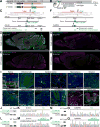
Figure 1. HDR-Mediated Genome Editing in the Embryonic Brain
(A) Schematic of AAV-HDR for the delivery of a sgRNA and HDR donor template to insert the HA tag or mEGFP sequence. U6, human U6 Polymerase III promoter. (B) Schematic illustration of experiments. (C and F) Graphical representation of the mouse genomic loci of β-Actin (C) and CaMKIIα (F) showing the target sites for Cas9. The sgRNA targeting sequences are labeled in magenta. The protospacer-adjacent motif (PAM) sequences are labeled in blue. The start codons of β-Actin and CaMKIIα are marked in orange. The Cas9 cleavage sites are indicated by the black arrowheads. PCR primer sets (control and recombination) for PCR genotyping (J, L, N, and P) are indicated by the arrows. (D, E, G, and H) Confocal microscopic images of whole sagittal brain sections of Cas9 (D and G) and wild type (E and H) mice showing immunoreactivities for NeuN (magenta) and the HA tag (green) fused to the N-terminus of endogenous β-Actin (D and E) and CaMKIIα (G and H). (I) Confocal microscopic images of various brain regions showing the DAPI signal (blue), mEGFP-CaMKIIα fluorescence (green), and immunoreactivities for NeuN, parvalbumin (PV), or tyrosine hydroxylase (TH) (magenta) and the HA tag (green) fused to the N-terminus of endogenous _β_-Actin or CaMKIIα. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer (Retina). DG, dentate gyrus (Hippocampus). HIP, hippocampus; v3, third ventricle (Thalamus). VTA, ventral tegmental area; white arrows, TH positive neurons (Midbrain). (J, L, N, and P) PCR genotyping using genomic DNAs extracted from the AAV-HDR injected brains of wild type and Cas9 mice. Recombination and control primer sets for HA-β-Actin (J), mEGFP-β-Actin (L), HA-CaMKIIα (N), and mEGFP-CaMKIIα (P) were used for PCR. (K, M, O, and Q) DNA sequencing analysis of the PCR products for HA-β-Actin (K), mEGFP-β-Actin (M), HA-CaMKIIα (O), and mEGFP-CaMKIIα (Q). The HA tag and mEGFP sequences are marked in light green and green, respectively. Scale bars, 1mm (D and G); 50 μm (I). See also Figures S1–4, STAR Methods, and Table S2 and S3.
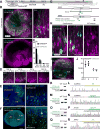
Figure 2. HDR-Mediated Genome Editing in Postmitotic Neurons in Organotypic Slice Cultures
(A) Schematic illustration of experiments. (B) Confocal microscopic images of organotypic hippocampal slice cultures of Cas9 (top) and wild type (bottom) mice transduced with AAV-HDR for HA-CaMKIIα using serotype 1, showing the EGFP fluorescence (magenta) and immunoreactivity for the HA tag (green) fused to the N-terminus of endogenous CaMKIIα. (C and D) The effect of the copy number of AAV-HDR on HA-CaMKIIα knockin efficiency. Black and gray bars, knockin efficiency in infected cells or all neurons in CA1, respectively (the ratio of the number of HA-positive cells to that of EGFP-positive or NeuN-positive cells, respectively; 100%, n (cell) = 497 (EGFP)/823 (NeuN); 50%, n = 345/553; 25%, n = 403/602; 12.5%, n = 271/477; 3 slices for each). Representative images of slice cultures transduced with different copy number of AAV-HDR for HA-CaMKIIα are shown (D). (E and F) Confocal microscopic images of organotypic hippocampal cultures prepared from Cas9 mice showing DAPI signal (blue) and mEGFP fluorescence (green) fused to the N-terminus of endogenous CaMKIIα (E) and β-Actin (F). Enlarged images of the white dotted squares in CA3 (E, left) and CA1 (F, left) were shown (E, right; F, right). (G) Graphical representation of the mouse genomic loci of ERK2 showing the target sites for Cas9. The sgRNA targeting sequences are labeled in magenta. The PAM sequence is labeled in blue. The start codons of ERK2 is marked in orange. The Cas9 cleavage sites are indicated by the black arrowheads. PCR primer sets (control and recombination) for PCR genotyping (Q) are indicated by the arrows. (H) Confocal microscopic images of organotypic hippocampal slice cultures of Cas9 (left and middle) and wild type (right) mice showing the EGFP fluorescence (magenta) and immunoreactivity for the HA tag (green) fused to the N-terminus of endogenous ERK2. The representative image of slice treated with 50 μM bicuculline for 1 h is shown (middle). (I) Confocal microscopic images of organotypic hippocampal slice cultures of Cas9 mice transduced with AAV-HDR for HA-CaMKIIα using serotype 9 showing the EGFP fluorescence (magenta) and immunoreactivity for the HA-CaMKIIα (green). (J) The effect of AAV serotypes on HA-CaMKIIα knockin efficiency. Black and gray squares, the knockin efficiency of AAV9-based AAV-HDR in infected cells or all neurons in CA1, respectively (the ratio of the number of HA-positive cells to that of EGFP-positive or NeuNpositive cells, respectively, n (cell) = 330 (EGFP)/604 (NeuN)/3 slices). Black and gray circles, the knockin efficiency of AAV1-based AAV-HDR (corresponding to 100% in Figure 2C) is also shown for comparison. The copy number of AAV9-based AAV-HDR is same as that of AAV1-based AAV-HDR (5.4×109 GC). (K, M, O, and Q) PCR genotyping using genomic DNA extracted from the AAV-transduced slices of wild type and Cas9 mice. Recombination and control primer sets for HA-CaMKIIα (K), mEGFP-CaMKIIα (M), mEGFP-β-Actin (O), HA-ERK2 (Q) were used for PCR. (L, N, P, and R) DNA sequencing analysis of the PCR products for HA-CaMKIIα (L), mEGFP-CaMKIIα (N), mEGFP-β-Actin (P), and HA-ERK2 (R). The HA or mEGFP tag sequences are marked in light green and green, respectively. Data are represented as mean ± SEM. Scale bars, 50 μm (B; right, D, E; right, F; right, H); 1 mm (B; left, I); 500 μm (E; left, F; left). See also Figures S5A–S5E and Table S2 and S3.
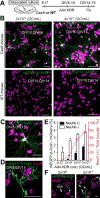
Figure 3. HDR-Mediated Genome Editing in Postmitotic Neurons in Primary Dissociated Cultures
(A) Schematic illustration of experiments. (B) Confocal microscopic images of dissociated hippocampal cultures prepared from Cas9 mice (top) and wild type mice (bottom) showing immunoreactivity for NeuN (magenta) and the HA tag (green) fused to the N-terminus of endogenous β-Actin (left; AAV-HDR, 2×1010 GC/ml) and CaMKIIα (right; AAV-HDR, 4×1010 GC/ml). Red arrows, NeuN positive neurons; white arrows, NeuN negative cells. (C and D) Enlarged images of mEGFP-β-Actin positive neuron (C) and astrocyte (D). (E and F) The effect of AAV-HDR concentration on mEGFP-β-Actin knockin efficiency in neurons and non-neuronal cells. Black and white bars, knockin efficiency of mEGFP-β-Actin in neurons and non-neuronal cells, respectively (the ratio of the number of mEGFP-positive and NeuN-positive or negative cells to that of mCherry-positive and NeuN-positive or negative cells, respectively; 2×1010, n (cell) = 1544/294 (mCherry and NeuN double-positive cells/mCherry-positive, NeuN-negative cells; 2×109, n = 968/203; 2×108, n = 1096/211; 2×107, n = 1276/222 from 4–6 fields of view in each of independent experiments). Red line, the fraction of neurons in total mEGFP-β-Actin knockin cells (the ratio of the number of mEGFP/NeuN double positive cells to that of total mEGFP-positive cells). Representative images of dissociated cultures transduced with AAV-HDR for mEGFP-β-Actin at a concentration of 2×108 and 2×1010 GC/ml are shown (F). Red arrows, NeuN positive neurons; white arrows, NeuN negative cells. Data are represented as mean ± SEM. Scale bars, 50 μm (B, C, D, and F).
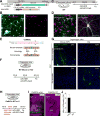
Figure 4. Dual AAV System for HDR-Mediated Genome Editing In Vitro
(A) Schematic of AAV-EFS-Cas9 and AAV-HDR. EFS, elongation factor-1 short promoter; Myc; c-Myc tag; NLS, nuclear localization signal; spA, synthetic polyadenylation signal. (B) Confocal microscopic images of hippocampal dissociated neurons showing the EGFP fluorescence (green) and immunoreactivity for the Myc tag (magenta) fused to the N-terminus of SpCas9. (C, F, and H) Schematic illustrations of experiments. (E) Genomic sequence comparisons of mouse and rat CaMKIIα. The sgRNA targeting sequences (magenta), PAM sequences (blue), and Cas9 cleavage sites (black arrowheads) are shown (top). Schematics of mouse and rat HDR template for HA-CaMKIIα (bottom) showing homology arms (orange) and HA (green). (G) Confocal microscopic images of organotypic hippocampal cultures prepared from wild type mice (left) and rats (right) showing DAPI signal (blue) and mEGFP fluorescence (green) fused to the N-terminus of endogenous CaMKIIα. (I) Confocal microscopic images of organotypic hippocampal slice cultures prepared from wild type mice (left) or CaMKIIα conditional knockout mice (right) showing the tdTomato fluorescence (magenta) and immunoreactivity for the HA tag (green) fused to the N-terminus of endogenous CaMKIIα. (J) The fraction of HA-CaMKIIα-positive cells in wild type and CaMKIIα deleted cells (the ratio of the number of HA-CaMKIIα-positive cells to that of tdTomato-positive cells). Wild-type, n = 375 cells, 3 slices; floxed, n = 374 cells, 3 slices. Data are represented as mean ± SEM. Scale bars, 50 μm (B, D, G, and I). See also Figures S5F–S5H.
Figure 5. Single and Dual AAV Systems for HDR-Mediated Genome Editing In Vivo
(A and G) Schematic illustrations of experiments. (B, D, H, J, and L) Schematic illustrations of AAV injection sites. (C, E, F, I, K, and M) Confocal microscopic images of coronal brain sections of the cerebral cortex (C and I), hippocampus (E and F), amygdala (K), and striatum (M) of Cas9 (C; left, E, and F) or wild type mice (C; right, I, K, and M) showing DAPI signal (blue), the EGFP fluorescence (magenta), and immunoreactivities for NeuN (blue) and the HA tag (green) fused to the N-terminus of endogenous CaMKIIα. Images of the cerebral cortex transduced with AAV-HDR and AAV-EFS-Cas9 (I, left) and AAV-HDR only (I, right) are shown. BLA, basolateral amygdala; CPu, caudate putamen; NAc, nucleus accumbens. Scale bars, 50 μ,m (C, F, I, K; right, M: right); 1 mm (E, K; left, and M; left). See also Figure S6.
Figure 6. HDR-Mediated Genome Editing in Aged, Disease-Model Mice
(A) Schematic illustration of experiments. (B–E) Confocal microscopic images of coronal brain sections of the CA1 hippocampus of wild type (B and D) and littermate J20 (C and E) mice at 1 year old showing the DAPI signal (blue), mCherry fluorescence (magenta), and mEGFP fluorescence (green) fused to the N-terminus of endogenous β-Actin. Representative images of the apical secondary dendrites of CA1 pyramidal neurons of wild type (D) and J20 (E) mice are shown. (F) The averaged density of spines on apical secondary dendrites of wild type (n = 946 spines/8 neurons) and J20 (n = 1001/9) mice. *p < 0.05, Student’s t test. Data are represented as mean ± SEM. Scale bars, 50 μm (B and C); 5 μm (D and E).
Similar articles
- Genome editing in the mammalian brain using the CRISPR-Cas system.
Nishiyama J. Nishiyama J. Neurosci Res. 2019 Apr;141:4-12. doi: 10.1016/j.neures.2018.07.003. Epub 2018 Aug 1. Neurosci Res. 2019. PMID: 30076877 Review. - Enhancing CRISPR/Cas9-mediated homology-directed repair in mammalian cells by expressing Saccharomyces cerevisiae Rad52.
Shao S, Ren C, Liu Z, Bai Y, Chen Z, Wei Z, Wang X, Zhang Z, Xu K. Shao S, et al. Int J Biochem Cell Biol. 2017 Nov;92:43-52. doi: 10.1016/j.biocel.2017.09.012. Epub 2017 Sep 18. Int J Biochem Cell Biol. 2017. PMID: 28928041 - Methods for In Vivo CRISPR/Cas Editing of the Adult Murine Retina.
Hung SS, Li F, Wang JH, King AE, Bui BV, Liu GS, Hewitt AW. Hung SS, et al. Methods Mol Biol. 2018;1715:113-133. doi: 10.1007/978-1-4939-7522-8_9. Methods Mol Biol. 2018. PMID: 29188510 - Enhancement of homology-directed repair with chromatin donor templates in cells.
Cruz-Becerra G, Kadonaga JT. Cruz-Becerra G, et al. Elife. 2020 Apr 28;9:e55780. doi: 10.7554/eLife.55780. Elife. 2020. PMID: 32343230 Free PMC article. - CRISPR/Cas9-mediated correction of human genetic disease.
Men K, Duan X, He Z, Yang Y, Yao S, Wei Y. Men K, et al. Sci China Life Sci. 2017 May;60(5):447-457. doi: 10.1007/s11427-017-9032-4. Epub 2017 May 3. Sci China Life Sci. 2017. PMID: 28534256 Review.
Cited by
- Start codon disruption with CRISPR/Cas9 prevents murine Fuchs' endothelial corneal dystrophy.
Uehara H, Zhang X, Pereira F, Narendran S, Choi S, Bhuvanagiri S, Liu J, Ravi Kumar S, Bohner A, Carroll L, Archer B, Zhang Y, Liu W, Gao G, Ambati J, Jun AS, Ambati BK. Uehara H, et al. Elife. 2021 Jun 8;10:e55637. doi: 10.7554/eLife.55637. Elife. 2021. PMID: 34100716 Free PMC article. - Current Status of Gene Therapy Research in Polyglutamine Spinocerebellar Ataxias.
Afonso-Reis R, Afonso IT, Nóbrega C. Afonso-Reis R, et al. Int J Mol Sci. 2021 Apr 19;22(8):4249. doi: 10.3390/ijms22084249. Int J Mol Sci. 2021. PMID: 33921915 Free PMC article. Review. - In vivo Genome Editing Therapeutic Approaches for Neurological Disorders: Where Are We in the Translational Pipeline?
Lubroth P, Colasante G, Lignani G. Lubroth P, et al. Front Neurosci. 2021 Feb 18;15:632522. doi: 10.3389/fnins.2021.632522. eCollection 2021. Front Neurosci. 2021. PMID: 33679313 Free PMC article. Review. - Building CRISPR Gene Therapies for the Central Nervous System: A Review.
Salomonsson SE, Clelland CD. Salomonsson SE, et al. JAMA Neurol. 2024 Mar 1;81(3):283-290. doi: 10.1001/jamaneurol.2023.4983. JAMA Neurol. 2024. PMID: 38285472 Review. - Next Generation Precision Medicine: CRISPR-mediated Genome Editing for the Treatment of Neurodegenerative Disorders.
Raikwar SP, Kikkeri NS, Sakuru R, Saeed D, Zahoor H, Premkumar K, Mentor S, Thangavel R, Dubova I, Ahmed ME, Selvakumar GP, Kempuraj D, Zaheer S, Iyer SS, Zaheer A. Raikwar SP, et al. J Neuroimmune Pharmacol. 2019 Dec;14(4):608-641. doi: 10.1007/s11481-019-09849-y. Epub 2019 Apr 23. J Neuroimmune Pharmacol. 2019. PMID: 31011884 Free PMC article. Review.
References
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous