A short linear motif in scaffold Nup145C connects Y-complex with pre-assembled outer ring Nup82 complex - PubMed (original) (raw)
A short linear motif in scaffold Nup145C connects Y-complex with pre-assembled outer ring Nup82 complex
Roman Teimer et al. Nat Commun. 2017.
Abstract
Nucleocytoplasmic transport occurs through nuclear pore complexes (NPCs), which are formed from multiple copies of ~30 different nucleoporins (Nups) and inserted into the double nuclear membrane. Many of these Nups are organized into subcomplexes, of which the Y-shaped Nup84 complex is the major constituent of the nuclear and cytoplasmic rings. The Nup82-Nup159-Nsp1 complex is another module that, however, is only assembled into the cytoplasmic ring. By means of crosslinking mass spectrometry, biochemical reconstitution, and molecular modeling, we identified a short linear motif in the unstructured N-terminal region of Chaetomium thermophilum Nup145C, a subunit of the Y-complex, that is sufficient to recruit the Nup82 complex, but only in its assembled state. This finding points to a more general mechanism that short linear motifs in structural Nups can act as sensors to cooperatively connect pre-assembled NPC modules, thereby facilitating the formation and regulation of the higher-order NPC assembly.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Fig. 1
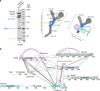
XL-MS analysis of the _Ct_Nup82 complex with the central vertex of the Y-shaped _Ct_Nup84 complex. a In vitro reconstitution of the _Ct_Nup82–Y complex (_Ct_Nup82–Nup159C–Nsp1C–Nup145C–Nup85–Nup120). An immobilized Flag-_Ct_Nup159C–Nup82–Nsp1C complex was incubated with the soluble _Ct_Nup145C–Nup85–Nup120 complex (Y-vertex). Shown is the analysis of the Flag eluate by SDS-PAGE and coomassie staining. _Ct_Nup145C is highlighted in blue as it interacts directly with the _Ct_Nup82 complex. M marker, MW molecular weight. An uncropped image of the gel is shown in Supplementary Fig. 4a. b Schematic representation of the _Ct_Nup82–Y complex showing the crosslinks determined by mass spectrometry. Crosslinks are depicted as straight gray lines, intramolecular self-links as curved purple lines, and intermolecular self-links as dark red loops. DIM domain invasion motif. Additional data are shown in Supplementary Table 2 and Supplementary Data 1
Fig. 2
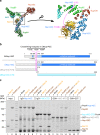
The N-terminal domain (NTD) of _Ct_Nup145C is both necessary and sufficient for the recruitment of the _Ct_Nup82 complex. a Crystal structure of the majority of the Y-shaped _Sc_Nup84 complex (PDB ID: 4xmm). The region upstream of the _Sc_Nup145C-DIM domain is indicated by a dashed pink line (right panel) as it is missing from the crystal structure. b Schematic representation of _Sc_Nup145C and _Ct_Nup145C and the design of truncated _Ct_Nup145C-NTD and _Ct_Nup145C-CTD; see also Supplementary Fig. 2. The crosslinking hotspots found in _Ct_Nup145C-NTD (Fig. 1b) are indicated by dashed pink lines. DIM domain invasion motif. c In vitro binding assay with immobilized _Ct_Nup145C, _Ct_Nup145C-NTD, or _Ct_Nup145C-CTD and soluble _Ct_Sec13 and/or the _Ct_Nup82–Nup159C–Nsp1C complex (Nup82-com). Shown is the analysis of SDS eluates by SDS-PAGE and coomassie staining. The experiment was performed at least twice with consistent results. Asterisks indicate bands corresponding to IgG heavy and light chains; M marker, Mock purification buffer with E. coli whole cell lysate, MW molecular weight. An uncropped image of the gel is shown in Supplementary Fig. 4b
Fig. 3
Interaction between the _Ct_Nup82 complex and the _Ct_Y-vertex. In vitro binding assay with the immobilized _Ct_Nup82 complex (Flag-_Ct_Nup159C–Nup82–Nsp1C) and different soluble prey proteins of the _Ct_Y-shaped complex. Shown is the analysis of SDS eluates by SDS-PAGE and coomassie staining. The experiment was performed at least twice with consistent results. The asterisk indicates bands corresponding to the anti-Flag light chain; M marker, Mock purification buffer with E. coli whole cell lysate, MW molecular weight, Y-vertex co-purified _Ct_Nup145C–Nup85–Nup120 complex, Y-vertex-CTD co-purified _Ct_Nup145C-CTD–Nup85–Nup120 complex. Uncropped images of the gels are shown in Supplementary Fig. 4c, d
Fig. 4
Identification of a short linear motif (SLiM) in _Ct_Nup145C-NTD that recruits the _Ct_Nup82 complex. a Schematic representation of _Sc_Nup145C and _Ct_Nup145C and the design of truncated _Ct_Nup145C-A and _Ct_Nup145C-B motifs; see also Supplementary Fig. 2. The crosslinking hotspots found in _Ct_Nup145C-NTD (Fig. 1b) are indicated by dashed pink lines. DIM domain invasion motif. b In vitro binding assay with Flag-immobilized _Ct_Nup145C-A, _Ct_Nup145C-B, and _Ct_Nup82 complex (Flag-_Ct_Nup159C–Nup82–Nsp1C) and soluble GST-_Ct_Nup145C-A, GST-_Ct_Nup145C-B, _Ct_Nup82 complex, and/or _Ct_Nup145N-ΔFG. Shown is the analysis of SDS eluates by SDS-PAGE and coomassie staining. The image section marked by the dashed box was subjected to image processing to increase the visibility of the small bait peptides _Ct_Nup145C-A-Flag and _Ct_Nup145C-B-Flag. c In vitro binding assay with Flag-immobilized _Ct_Nsp1C, _Ct_Nup159C, _Ct_Nup82, and _Ct_Nup82 complex (Flag-_Ct_Nup159C–Nup82–Nsp1C) and soluble GST-_Ct_Nup145C-B or _Ct_Nup145C. The experiments were performed at least twice with consistent results. The asterisk indicates bands corresponding to the anti-Flag light chain; M marker, Mock purification buffer with E. coli whole cell lysate, MW molecular weight. Uncropped gel images are shown in Supplementary Fig. 4e, f
Fig. 5
XL-MS suggests that _Ct_Nup145C binding to the _Ct_Nup82 complex resembles _Ct_Nic96 binding to the _Ct_Nsp1 complex. The model of the _Ct_Nup82 complex a was built based on the _Ct_Nsp1 complex (b, PDB ID: 5cws). The _Ct_Nup145C-82CIM motif (residues 215–270), which could not be reliably modeled at the atomic level, is shown as a low-resolution density. Note that although sequence similarity suggests that the _Ct_Nup82 complex has an architecture similar to the _Ct_Nsp1 complex; the exact orientation of the coiled-coil domains might be different (curved double arrows). Residues crosslinked to _Ct_Nup145C-82CIM are shown as red spheres and the crosslinked residues of _Ct_Nup145C are marked as K242, K258, and K266. Crosslinks between _Ct_Nup82, _Ct_Nsp1, and _Ct_Nup159 are indicated as sticks, crosslinks to _Ct_Nup145C as dashed lines. Crosslinks that satisfy the distance threshold of 30 Å are colored blue, and red otherwise. The two violated (red) crosslinks (_Ct_Nup159 Lys1249-_Ct_Nup82 Lys710 and _Ct_Nsp1 Lys649-_Ct_Nup82 Lys769) have relatively low ld-scores of 25.3 and 28.46, respectively. They thus might be false positives, in line with the calibrated FDR of 5% for the entire XL-MS data set. Alternatively, flexibility or alternative arrangements of coiled-coil domains might account for the distance violations
Similar articles
- Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel.
Fischer J, Teimer R, Amlacher S, Kunze R, Hurt E. Fischer J, et al. Nat Struct Mol Biol. 2015 Oct;22(10):774-81. doi: 10.1038/nsmb.3084. Epub 2015 Sep 7. Nat Struct Mol Biol. 2015. PMID: 26344569 - Architecture of the cytoplasmic face of the nuclear pore.
Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, Brown B, Tang AW, Rundlet EJ, Correia AR, Chen S, Regmi SG, Stevens TA, Jette CA, Dasso M, Patke A, Palazzo AF, Kossiakoff AA, Hoelz A. Bley CJ, et al. Science. 2022 Jun 10;376(6598):eabm9129. doi: 10.1126/science.abm9129. Epub 2022 Jun 10. Science. 2022. PMID: 35679405 Free PMC article. - Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile.
Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Amlacher S, et al. Cell. 2011 Jul 22;146(2):277-89. doi: 10.1016/j.cell.2011.06.039. Cell. 2011. PMID: 21784248 - The nuclear pore complex: understanding its function through structural insight.
Beck M, Hurt E. Beck M, et al. Nat Rev Mol Cell Biol. 2017 Feb;18(2):73-89. doi: 10.1038/nrm.2016.147. Epub 2016 Dec 21. Nat Rev Mol Cell Biol. 2017. PMID: 27999437 Review. - Structure and Assembly of the Nuclear Pore Complex.
Hampoelz B, Andres-Pons A, Kastritis P, Beck M. Hampoelz B, et al. Annu Rev Biophys. 2019 May 6;48:515-536. doi: 10.1146/annurev-biophys-052118-115308. Epub 2019 Apr 3. Annu Rev Biophys. 2019. PMID: 30943044 Review.
Cited by
- Fantastic nuclear envelope herniations and where to find them.
Thaller DJ, Patrick Lusk C. Thaller DJ, et al. Biochem Soc Trans. 2018 Aug 20;46(4):877-889. doi: 10.1042/BST20170442. Epub 2018 Jul 19. Biochem Soc Trans. 2018. PMID: 30026368 Free PMC article. Review. - Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis.
Huang G, Zhang Y, Zhu X, Zeng C, Wang Q, Zhou Q, Tao Q, Liu M, Lei J, Yan C, Shi Y. Huang G, et al. Cell Res. 2020 Jun;30(6):520-531. doi: 10.1038/s41422-020-0319-4. Epub 2020 May 6. Cell Res. 2020. PMID: 32376910 Free PMC article. - Exploring Cellular Gateways: Unraveling the Secrets of Disordered Proteins within Live Nuclear Pores.
Yu W, Tingey M, Kelich JM, Li Y, Yu J, Junod SL, Jiang Z, Hansen I, Good N, Yang W. Yu W, et al. Res Sq [Preprint]. 2024 Jan 9:rs.3.rs-3504130. doi: 10.21203/rs.3.rs-3504130/v1. Res Sq. 2024. PMID: 38260360 Free PMC article. Preprint. - The First MS-Cleavable, Photo-Thiol-Reactive Cross-Linker for Protein Structural Studies.
Iacobucci C, Piotrowski C, Rehkamp A, Ihling CH, Sinz A. Iacobucci C, et al. J Am Soc Mass Spectrom. 2019 Jan;30(1):139-148. doi: 10.1007/s13361-018-1952-8. Epub 2018 Apr 20. J Am Soc Mass Spectrom. 2019. PMID: 29679287 - One Ring to Rule them All? Structural and Functional Diversity in the Nuclear Pore Complex.
Fernandez-Martinez J, Rout MP. Fernandez-Martinez J, et al. Trends Biochem Sci. 2021 Jul;46(7):595-607. doi: 10.1016/j.tibs.2021.01.003. Epub 2021 Feb 6. Trends Biochem Sci. 2021. PMID: 33563541 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources