Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution - PubMed (original) (raw)
Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution
Nicholas McGranahan et al. Cell. 2017.
Abstract
Immune evasion is a hallmark of cancer. Losing the ability to present neoantigens through human leukocyte antigen (HLA) loss may facilitate immune evasion. However, the polymorphic nature of the locus has precluded accurate HLA copy-number analysis. Here, we present loss of heterozygosity in human leukocyte antigen (LOHHLA), a computational tool to determine HLA allele-specific copy number from sequencing data. Using LOHHLA, we find that HLA LOH occurs in 40% of non-small-cell lung cancers (NSCLCs) and is associated with a high subclonal neoantigen burden, APOBEC-mediated mutagenesis, upregulation of cytolytic activity, and PD-L1 positivity. The focal nature of HLA LOH alterations, their subclonal frequencies, enrichment in metastatic sites, and occurrence as parallel events suggests that HLA LOH is an immune escape mechanism that is subject to strong microenvironmental selection pressures later in tumor evolution. Characterizing HLA LOH with LOHHLA refines neoantigen prediction and may have implications for our understanding of resistance mechanisms and immunotherapeutic approaches targeting neoantigens. VIDEO ABSTRACT.
Keywords: bioinformatics; cancer evolution; chromosomal instability; copy number; heterogeneity; immune-editing; immune-escape; loss of heterozygosity; lung cancer; neoantigen.
Copyright © 2017 The Francis Crick Institute. Published by Elsevier Inc. All rights reserved.
Figures
Graphical abstract
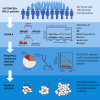
Figure 1
Outline and Validation of LOHHLA for Inference of HLA Class I Allele-Specific Copy Number in Tumors (A) Schematic of the LOHHLA algorithm. (B) Comparison of minor allele copy number for ASCAT and LOHHLA. (C) Venn diagram illustrating LOHHLA and ASCAT comparison for inference of allelic imbalance at HLA locus. (D) Venn diagram illustrating LOHHLA and ASCAT comparison for inference of LOH at HLA locus. See also Figures S1 and S2.
Figure S1
Comparison of LOHHLA and ASCAT, Related to Figure 1 (A) Plot illustrating comparison of ASCAT major copy number and LOHHLA major copy number. (B and C) Summary of concordant and discordant tumor regions in terms of allelic imbalance (B) and LOH (C). (D) Schematic illustrating how ASCAT cannot directly infer HLA copy number or which HLA allele is subject to loss. By contrast, LOHHLA uses SNPs covering HLA genes to directly infer HLA copy number.
Figure S2
Validation of LOHHLA Using Fragment Analysis, Related to Figure 1 (A) Area under the curve of each allele using the Applied Biosystems software GeneMapper v5 for germline and tumor regions R1 and R2 in CRUK0010. (B) Normalized allelic ratio determined using the formula (Atumor/Btumor)/(Anormal/Bnormal). Notably, region R1 shows clear evidence of allelic imbalance and likely LOH, while region R2 appears similar to germline. (C–H) Normalized allelic ratio for tumor regions showing either LOH and allelic imbalance; no LOH but allelic imbalance; or no LOH or allelic imbalance classified by LOHHLA (C), ASCAT (D), TITAN (E and G) and Sequenza (F and H). Tumor purity, as assessed by ASCAT is depicted for each tumor region, p values correspond to Wilcoxon rank sum test.
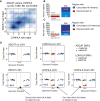
Figure 2
Frequency and Timing of HLA LOH in NSCLC (A) The total number of lung adenocarcinoma and lung squamous cell carcinoma TRACERx patients exhibiting an HLA non-synonymous mutation, HLA allelic imbalance (AI), or LOH at the HLA locus is shown. (B) Proportion of HLA allelic imbalance (AI) and HLA LOH identified in NSCLC by sub-type. Enrichment significance was tested using a Fisher’s exact test. (C and D) Pie charts show the timing of HLA LOH events using multi-region information for lung adenocarcinoma (C) and lung squamous cell carcinomas (D). Events at individual HLA A/B/C loci were considered clonal if they were found in every region considered and subclonal if they were found in only a subset of tumor regions. A patient sample was considered to have clonal HLA LOH if all of the individual loci lost in that tumor occurred clonally. Two lung squamous cell carcinoma patients with only a single region available for copy-number analysis are not shown. (E and F) Phylogenetic trees for each lung adenocarcinoma (E) and lung squamous cell carcinomas (F) showing evidence of HLA LOH have been annotated with the most likely timing of the HLA LOH event. Homozygous HLA alleles, where HLA LOH is not possible, are indicated by an asterisk. Clones on the phylogenetic tree (nodes) are indicated as clonal (blue) or subclonal (red). In cases where the HLA LOH event did not map to a possible clone on the phylogenetic tree, an additional gray subclone was included. (G) Number of NSCLC patients from Brastianos et al. (2015) with paired primary/brain metastasis sequencing data available exhibiting no HLA LOH (gray), HLA LOH in both the primary tumor and brain metastasis (green), HLA LOH only in the primary tumor (red), or HLA LOH only in the brain metastasis (blue). Patients with HLA LOH identified consistently across HLA loci in both the primary tumor and every brain metastases were considered to have clonal HLA LOH. Patients with inconsistent HLA loci subject to LOH or those with HLA LOH identified in only a primary or brain metastasis sample were considered to have subclonal HLA LOH. (H) Timing of the HLA LOH events. Clonal HLA LOH events occur in both the primary tumor sample and the brain metastases (green), whereas subclonal HLA LOH events either arise in the brain metastases (blue) or have occurred in a subclone of the primary tumor that does not seed the brain metastasis (red). Overall, an increase in HLA LOH is observed in the brain metastases samples as compared to the primary tumor (27% to 43%) and a corresponding decrease is observed in brain metastases samples exhibiting no HLA LOH (73% to 57%).
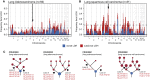
Figure 3
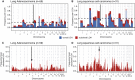
HLA LOH Reflects Selection in NSCLC (A and B) Frequency of focal LOH in lung adenocarcinoma (A) and lung squamous cell carcinoma (B). Focal LOH is defined as <75% of a chromosome arm. Arrow indicates location of HLA locus. Horizontal dashed line depicts significant focal LOH at p = 0.05, using simulations. Clonal LOH is shown in blue, with subclonal LOH shown in red. Chromosome arm LOH and focal subclonal LOH is shown in Figure S3. (C) Parallel evolution of HLA LOH, with allele-specific HLA loss shown on phylogenetic trees. See also Figure S3.
Figure S3
Arm-Level and Focal Subclonal LOH across the Genome, Related to Figure 3 (A and B) Arm-level LOH across the genome for lung adenocarcinoma (A) and lung squamous cell carcinoma (B). Arm-level LOH is defined as > 75% of a chromosome arm. Arrow indicates location of HLA locus. Horizontal dashed line depicts significant focal LOH at p = 0.05, using simulations. Clonal LOH is shown in blue, with subclonal LOH shown in red. (C and D) Focal subclonal LOH across the genome for lung adenocarcinoma (C) and lung squamous cell carcinoma (D). Focal LOH is defined as < 75% of a chromosome arm. Arrow indicates location of HLA locus.
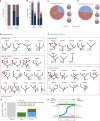
Figure 4
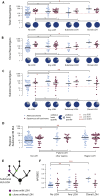
Non-synonymous Mutational Burden Associates with HLA LOH, and Neoantigens More Frequently Bind the Lost Allele (A) The total number of nonsynonymous mutations is plotted across different categories of HLA LOH for lung adenocarcinoma (light blue) and lung squamous cell carcinomas (magenta). Tumors were classified as having: no HLA LOH; any HLA LOH event, without taking into account the timing of the event; or clonal HLA LOH. The lowest total non-synonymous mutation quartile is indicated by the dashed red line and the proportion of tumors with a total non-synonymous mutational burden greater or less than that is indicated by the pie charts for each HLA LOH classification group. (B) The number of clonal non-synonymous mutations is plotted across different categories of HLA LOH for lung adenocarcinoma (light blue) and lung squamous cell carcinomas (magenta). (C) The number of subclonal non-synonymous mutations is plotted across different categories of HLA LOH for lung adenocarcinoma (light blue) and lung squamous cell carcinomas (magenta). All p values are calculated using an unpaired Wilcoxon test. (D) The number of non-synonymous mutations found in the clone harboring the HLA LOH event compared to the number of non-synonymous mutations in its sister clone, descended from the same ancestral cancer cell, but without HLA LOH. The p value is calculated using a paired Wilcoxon test. (E) The number of subclonal neoantigens predicted to bind to either the lost HLA allele or the kept HLA allele is indicated for all NSCLC tumors exhibiting HLA LOH, all lung adenocarcinoma tumors with HLA LOH, and all lung squamous tumors with HLA LOH. A red line indicates an elevated subclonal neoantigen mutation burden in the HLA LOH subclone compared to the subclone without HLA LOH, while blue indicates the converse. The p value is calculated using a paired Wilcoxon test. (F) The total number of mutations predicted to result in a binder to the lost allele is shown for all patients with at least one HLA LOH event. The mutation clonality is also indicated as either clonal (light blue) or subclonal (light red). See also Figure S4.
Figure S4
Neoantigen and Regional HLA LOH Associations, Related to Figure 4 (A) The total number of neoantigens is plotted across different categories of HLA LOH for lung adenocarcinoma (light blue) and lung squamous cell carcinomas (magenta). Tumors were classified as having: no HLA LOH; any HLA LOH event, without taking into account the timing of the event; subclonal HLA LOH; or clonal HLA LOH. The lowest total neoantigen quartile is indicated by the dashed red line and the proportion of tumors with a total neoantigen burden greater or less than that is indicated by the pie charts for each HLA LOH classification group. (B) The number of clonal neoantigens is plotted across different categories of HLA LOH for lung adenocarcinoma (light blue) and lung squamous cell carcinomas (magenta). (C) The number of subclonal neoantigens is plotted across different categories of HLA LOH for lung adenocarcinoma (light blue) and lung squamous cell carcinomas (magenta). (D) The number of subclonal non-synonymous mutations is plotted for tumor regions from tumors without any indication of HLA LOH, for tumor regions without HLA LOH from a tumor with other regions harboring HLA LOH, and for tumor regions containing an HLA LOH event. All p values are calculated using an unpaired wilcoxon test. (E) Schematic of the clones considered for the comparison performed in Figure 4D. Here, the cancer subclone harboring HLA loss (purple) is shown with its sister subclone, descended from the same ancestral cancer cell, but without HLA loss (green). (F) For each lung adenocarcinoma (blue) and lung squamous cell carcinoma (purple) tumor, the relative contributions of APOBEC mutational signatures are shown. p values are calculated using an unpaired wilcoxon test.
Figure 5
HLA LOH and Immune Phenotypes (A) Anti-PD-L1 staining on FFPE diagnostic blocks from tumors with clonal HLA LOH, subclonal HLA LOH, and no observed HLA LOH. Immune-cell-based staining and tumor-cell staining is depicted. (B) Staining from two representative tumors, one without HLA LOH and one with clonal HLA LOH is shown. (C) The log-ratio of medians between tumors containing an HLA LOH event at all loci and those without any HLA LOH event is shown for published immune microenvironment measures and signatures. Increase of an immune measure among tumors with HLA LOH is shown in red, and a decrease is shown in blue. False discovery rate (FDR) (q) values comparing the distribution of immune measures between the HLA LOH groups are indicated by asterisks (∗). See also Figure S5 and Table S1.
Figure S5
Frequency and Association with Mutational Burden of HLA LOH in TCGA, Related to Figure 5 (A) The total number of TCGA patients exhibiting an allelic imbalance or LOH at the HLA locus is shown. (B) The total number of nonsynonymous mutations is plotted across different categories of HLA LOH for lung adenocarcinoma (light blue) and lung squamous cell carcinomas (magenta). Tumors were classified as having: no HLA LOH; any HLA LOH event; or HLA LOH at all three HLA loci. The lowest total non-synonymous mutation quartile is indicated by the dashed red line and the proportion of tumors with a total non-synonymous mutational burden greater or less than that is indicated by the pie charts for each HLA LOH classification group.
Figure 6
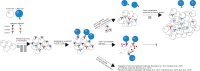
Model of HLA Allele-Specific Loss in NSCLC Model illustrating how HLA LOH may lead to immune escape in tumors. During tumor evolution, the accumulation of neoantigens may induce local immune infiltrates, including CD8 T cells. Local immune infiltrates may act as a selection barrier for tumors. Subclones with HLA LOH may be positively selected as these can evade killing by avoiding CD8 T cell recognition. Alternatively, other subclones may evade killing through other mechanisms.
Comment in
- Immunotherapy: Nice to see you - evolving MHC I-peptide presentation.
Romero D. Romero D. Nat Rev Clin Oncol. 2018 Jan;15(1):5. doi: 10.1038/nrclinonc.2017.180. Epub 2017 Nov 14. Nat Rev Clin Oncol. 2018. PMID: 29134958 No abstract available. - Tumor Evolution: A Problem of Histocompatibility.
Vokes NI, Van Allen EM. Vokes NI, et al. Cell. 2017 Nov 30;171(6):1252-1253. doi: 10.1016/j.cell.2017.11.012. Cell. 2017. PMID: 29195070 Free PMC article. - The evolving understanding of immunoediting and the clinical impact of immune escape.
McCoach CE, Bivona TG. McCoach CE, et al. J Thorac Dis. 2018 Mar;10(3):1248-1252. doi: 10.21037/jtd.2018.03.60. J Thorac Dis. 2018. PMID: 29708132 Free PMC article. No abstract available.
Similar articles
- Neoantigen-directed immune escape in lung cancer evolution.
Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT, Lund T, Tanić M, Reading JL, Joshi K, Henry JY, Ghorani E, Wilson GA, Birkbak NJ, Jamal-Hanjani M, Veeriah S, Szallasi Z, Loi S, Hellmann MD, Feber A, Chain B, Herrero J, Quezada SA, Demeulemeester J, Van Loo P, Beck S, McGranahan N, Swanton C; TRACERx consortium. Rosenthal R, et al. Nature. 2019 Mar;567(7749):479-485. doi: 10.1038/s41586-019-1032-7. Epub 2019 Mar 20. Nature. 2019. PMID: 30894752 Free PMC article. - Somatic HLA Class I Loss Is a Widespread Mechanism of Immune Evasion Which Refines the Use of Tumor Mutational Burden as a Biomarker of Checkpoint Inhibitor Response.
Montesion M, Murugesan K, Jin DX, Sharaf R, Sanchez N, Guria A, Minker M, Li G, Fisher V, Sokol ES, Pavlick DC, Moore JA, Braly A, Singal G, Fabrizio D, Comment LA, Rizvi NA, Alexander BM, Frampton GM, Hegde PS, Albacker LA. Montesion M, et al. Cancer Discov. 2021 Feb;11(2):282-292. doi: 10.1158/2159-8290.CD-20-0672. Epub 2020 Oct 30. Cancer Discov. 2021. PMID: 33127846 - The absence of HLA class I expression in non-small cell lung cancer correlates with the tumor tissue structure and the pattern of T cell infiltration.
Perea F, Bernal M, Sánchez-Palencia A, Carretero J, Torres C, Bayarri C, Gómez-Morales M, Garrido F, Ruiz-Cabello F. Perea F, et al. Int J Cancer. 2017 Feb 15;140(4):888-899. doi: 10.1002/ijc.30489. Epub 2016 Nov 7. Int J Cancer. 2017. PMID: 27785783 - Loss of Heterozygosity (LOH) Affecting HLA Genes in Breast Cancer: Clinical Relevance and Therapeutic Opportunities.
Garrido MA, Navarro-Ocón A, Ronco-Díaz V, Olea N, Aptsiauri N. Garrido MA, et al. Genes (Basel). 2024 Nov 28;15(12):1542. doi: 10.3390/genes15121542. Genes (Basel). 2024. PMID: 39766811 Free PMC article. Review. - Immunotherapy in non-small-cell lung cancer: a bridge between research and clinical practice.
Passiglia F, Commendatore O, Vitali M, Conca R. Passiglia F, et al. Future Oncol. 2018 Jun;14(13s):41-60. doi: 10.2217/fon-2018-0098. Future Oncol. 2018. PMID: 29989452 Review.
Cited by
- HLA RNA Sequencing With Unique Molecular Identifiers Reveals High Allele-Specific Variability in mRNA Expression.
Johansson T, Yohannes DA, Koskela S, Partanen J, Saavalainen P. Johansson T, et al. Front Immunol. 2021 Feb 25;12:629059. doi: 10.3389/fimmu.2021.629059. eCollection 2021. Front Immunol. 2021. PMID: 33717155 Free PMC article. - The immune escape signature predicts the prognosis and immunotherapy sensitivity for pancreatic ductal adenocarcinoma.
Lu H, Zheng LY, Wu LY, Chen J, Xu N, Mi SC. Lu H, et al. Front Oncol. 2022 Sep 6;12:978921. doi: 10.3389/fonc.2022.978921. eCollection 2022. Front Oncol. 2022. PMID: 36147906 Free PMC article. - Adoptive transfer of personalized neoantigen-reactive TCR-transduced T cells in metastatic colorectal cancer: phase 2 trial interim results.
Parkhurst M, Goff SL, Lowery FJ, Beyer RK, Halas H, Robbins PF, Prickett TD, Gartner JJ, Sindiri S, Krishna S, Zacharakis N, Ngo L, Ray S, Bera A, Shepherd R, Levin N, Kim SP, Copeland A, Nah S, Levi S, Parikh N, Kwong MLM, Klemen ND, Yang JC, Rosenberg SA. Parkhurst M, et al. Nat Med. 2024 Sep;30(9):2586-2595. doi: 10.1038/s41591-024-03109-0. Epub 2024 Jul 11. Nat Med. 2024. PMID: 38992129 Clinical Trial. - Converging and evolving immuno-genomic routes toward immune escape in breast cancer.
Blanco-Heredia J, Souza CA, Trincado JL, Gonzalez-Cao M, Gonçalves-Ribeiro S, Gil SR, Pravdyvets D, Cedeño S, Callari M, Marra A, Gazzo AM, Weigelt B, Pareja F, Vougiouklakis T, Jungbluth AA, Rosell R, Brander C, Tresserra F, Reis-Filho JS, Tiezzi DG, de la Iglesia N, Heyn H, De Mattos-Arruda L. Blanco-Heredia J, et al. Nat Commun. 2024 Feb 21;15(1):1302. doi: 10.1038/s41467-024-45292-1. Nat Commun. 2024. PMID: 38383522 Free PMC article. - Chromosomal Instability, Selection and Competition: Factors That Shape the Level of Karyotype Intra-Tumor Heterogeneity.
van den Bosch T, Derks S, Miedema DM. van den Bosch T, et al. Cancers (Basel). 2022 Oct 12;14(20):4986. doi: 10.3390/cancers14204986. Cancers (Basel). 2022. PMID: 36291770 Free PMC article. Review.
References
- Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., Australian Pancreatic Cancer Genome Initiative. ICGC Breast Cancer Consortium. ICGC MMML-Seq Consortium. ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. - PMC - PubMed
- Brastianos P.K., Carter S.L., Santagata S., Cahill D.P., Taylor-Weiner A., Jones R.T., Van Allen E.M., Lawrence M.S., Horowitz P.M., Cibulskis K. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5:1164–1177. - PMC - PubMed
- Campbell J.D., Alexandrov A., Kim J., Wala J., Berger A.H., Pedamallu C.S., Shukla S.A., Guo G., Brooks A.N., Murray B.A., Cancer Genome Atlas Research Network Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 2016;48:607–616. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- FC001202/WT_/Wellcome Trust/United Kingdom
- 15951/CRUK_/Cancer Research UK/United Kingdom
- 20466/CRUK_/Cancer Research UK/United Kingdom
- 20613/CRUK_/Cancer Research UK/United Kingdom
- MR/P024351/1/MRC_/Medical Research Council/United Kingdom
- 18892/CRUK_/Cancer Research UK/United Kingdom
- FC001169/WT_/Wellcome Trust/United Kingdom
- 19310/CRUK_/Cancer Research UK/United Kingdom
- 20465/CRUK_/Cancer Research UK/United Kingdom
- 21999/CRUK_/Cancer Research UK/United Kingdom
- 22795/CRUK_/Cancer Research UK/United Kingdom
- MC_UP_1203/1/MRC_/Medical Research Council/United Kingdom
- 23464/CRUK_/Cancer Research UK/United Kingdom
- 17786/CRUK_/Cancer Research UK/United Kingdom
- 20276/CRUK_/Cancer Research UK/United Kingdom
- 20275/CRUK_/Cancer Research UK/United Kingdom
- 19278/CRUK_/Cancer Research UK/United Kingdom
- 17891/CRUK_/Cancer Research UK/United Kingdom
- 13462/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials