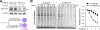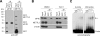m6A Facilitates eIF4F-Independent mRNA Translation - PubMed (original) (raw)
m6A Facilitates eIF4F-Independent mRNA Translation
Ryan A Coots et al. Mol Cell. 2017.
Abstract
In eukaryotic cells, protein synthesis typically begins with the binding of eIF4F to the 7-methylguanylate (m7G) cap found on the 5' end of the majority of mRNAs. Surprisingly, overall translational output remains robust under eIF4F inhibition. The broad spectrum of eIF4F-resistant translatomes is incompatible with cap-independent translation mediated by internal ribosome entry sites (IRESs). Here, we report that N6-methyladenosine (m6A) facilitates mRNA translation that is resistant to eIF4F inactivation. Depletion of the methyltransferase METTL3 selectively inhibits translation of mRNAs bearing 5' UTR methylation, but not mRNAs with 5' terminal oligopyrimidine (TOP) elements. We identify ABCF1 as a critical mediator of m6A-promoted translation under both stress and physiological conditions. Supporting the role of ABCF1 in m6A-facilitated mRNA translation, ABCF1-sensitive transcripts largely overlap with METTL3-dependent mRNA targets. By illustrating the scope and mechanism of eIF4F-independent mRNA translation, these findings reshape our current perceptions of cellular translational pathways.
Keywords: ABCF1; METTL3; cap-independent translation; eIF4F; epitranscriptome; mRNA methylation; stress response; translation.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
Figure 1. A Substantial Amount of Cellular Translation is Resistant to eIF4F Inhibition
(A) Sucrose gradient-based polysome profiling of eIF2α (S/S) and eIF2α (A/A) MEF cells before and after 1 h of amino acid starvation. The right panel shows the monosome/polysome ratio calculated using areas below the curve. Error bars, mean ± s.e.m.; n =3, biological replicates. (B) Global protein synthesis in starved eIF2α(S/S) and eIF2α(A/A) MEF cells was quantified from puromycin labeling. Error bars, mean ± s.e.m.; n =3, biological replicates. (C) Immunoblotting of mTORC1 downstream targets in eIF2α(S/S) and eIF2α(A/A) MEF cells before and after 1 h of amino acid starvation. (D) Immunoblotting of m7GTP pulldown assay in eIF2α(S/S) and eIF2α(A/A) MEF cells before and after 1 h of amino acid starvation. See also Figure S1 – S3.
Figure 2. Physiological Cap-Independent Translation is Dependent on m6A Modification
(A) Immunoblotting of MEFs with or without METTL3 knockdown. The lower panel shows a schematic diagram distinguishing cap-dependent and cap-independent translation. (B) Global protein synthesis in MEF cells with or without METTL3 knockdown was measured after pre-treatment of 1 µM Torin1 for various times. The right panel shows quantification of [35S] autoradiograph after Torin1 treatment. Error bars, mean ± s.e.m.; n =3, biological replicates. See also Figure S4.
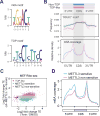
Figure 3. m6A Mediates eIF4F-Independent Translation
(A) Sequence logo representing the consensus motif relative to m6A (upper panel) and TOP elements (lower panel, derived from Thoreen et al, 2012). For m6A motif, “0” indicates the position of m6A. For TOP motif, “1” represents the 5’ end position. (B) The top panel shows the frequency of m6A motif across TOP-like (pink line) and non-TOP (blue line) mRNAs. The lower panel shows the distribution of m6A modification across TOP-like (pink line) and non-TOP (blue line) mRNAs using m6A-seq data sets obtained from MEFs. (C) Scatter plot shows the TE fold change in MEF cells in response to Torin1 treatment or METTL3 knockdown. Blue dots refer to TOP-like transcripts. Both METTL3-sensitive (bottom 10%) and non-sensitive mRNAs (top 10%) are highlighted. (D) m6A coverage obtained from m6A-seq was plotted for METTL3 non-sensitive (pink line) and METTL3 sensitive (blue line) mRNAs. Relative regions of 5’UTR, CDS, and 3’UTR are shown as the same size. See also Figure S4.
Figure 4. METTL3 Recognizes m6A but not the m7G Cap Structure
(A) Recombinant eIF4E and METTL3 proteins were purified from E. coli as GST fusion proteins followed by incubation with immobilized beads coated with m7GTP. Immunoblotting was conducted using an anti-GST antibody. (B) Whole cell lysates from HEK293 cells with or without Torin1 treatment were incubated with immobilized beads coated with m7GTP followed by immunoblotting using antibodies indicated. (C) Synthesized mRNA probes with or without single m6A were radiolabeled with 32P followed by incubation with an increasing dose of recombinant METTL3 (0, 0.3, 0.6, 1.2, 2.4 µg). The mRNA-protein complexes were resolved on a SDS-PAGE gel. See also Figure S5.
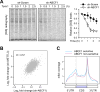
Figure 5. ABCF1 is Essential in Cap-Independent Translation of Hsp70
(A) The left panel shows the schematic of quantitative mass spectrometry using iTRAQ. Proteins enriched on β-actin mRNA (middle) or Hsp70 mRNA (right) purified from HeLa cells after heat shock stress are presented as scatter plots. The original peptide score (log2) and stress-induced fold changes (log2) are shown in the x-axis and the y-axis, respectively. (B) Whole cell lysates from heat shocked HeLa cells were subjected to immunoprecipitation using antibodies indicated followed by immunoblotting. (C) MEF cells with or without ABCF1 knockdown (Scramble) were collected at indicated times after heat shock stress (43°C, 1 h) followed by immunoblotting. N: no heat shock. See also Figure S6.
Figure 6. ABCF1 Mediates eIF4F-Independent Translation
(A) Global protein synthesis in MEF cells with or without ABCF1 knockdown was measured after pre-treatment of 1 µM Torin1 for various times. The right panel shows quantification of [35S] autoradiograph after Torin1 treatment. Error bars, mean ± s.e.m.; n =3, biological replicates. (B) Ribosome profiling data from cells with ABCF1 or METTL3 knockdown were used to determine the fold changes of TE. A scatter plot is presented to show positive correlation. (C) m6A coverage obtained from m6A-seq was plotted for ABCF1-sensitive (blue line) and non-responsive (pink line) mRNAs. Relative regions of 5’UTR, CDS, and 3’UTR are shown as the same size. See also Figure S6.
Figure 7. ABCF1 Mediates Translational Control of METTL3
(A) MEF cells with ABCF1 or METTL3 knockdown were treated with 5 µM MG132 for 16 hr. Whole cell lysates were collected for immunoblotting using antibodies indicated. (B) Total RNAs were purified from MEF cells with ABCF1 or METTL3 knockdown followed by qPCR. Error bars, mean ± s.e.m.; n =3, biological replicates. (C) m6A coverage of METTL3 mRNA using m6A-seq data sets obtained from HeLa and HEK293 cells. The transcript architecture is shown above. (D) MEF cells were transfected with Fluc plasmids shown in the left and Fluc levels were recorded by real time luminometry. Error bars, mean ± s.e.m.; n =3, biological replicates. *, p < 0.05 (t-test). See also Figure S7.
Similar articles
- A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent.
Terenin IM, Andreev DE, Dmitriev SE, Shatsky IN. Terenin IM, et al. Nucleic Acids Res. 2013 Feb 1;41(3):1807-16. doi: 10.1093/nar/gks1282. Epub 2012 Dec 24. Nucleic Acids Res. 2013. PMID: 23268449 Free PMC article. - Migration of Small Ribosomal Subunits on the 5' Untranslated Regions of Capped Messenger RNA.
Shirokikh NE, Dutikova YS, Staroverova MA, Hannan RD, Preiss T. Shirokikh NE, et al. Int J Mol Sci. 2019 Sep 10;20(18):4464. doi: 10.3390/ijms20184464. Int J Mol Sci. 2019. PMID: 31510048 Free PMC article. - A Cap for Every Occasion: Alternative eIF4F Complexes.
Ho JJD, Lee S. Ho JJD, et al. Trends Biochem Sci. 2016 Oct;41(10):821-823. doi: 10.1016/j.tibs.2016.05.009. Epub 2016 Jun 6. Trends Biochem Sci. 2016. PMID: 27283511 Free PMC article. Review. - Eukaryotic initiation factor (eIF) 4F binding to barley yellow dwarf virus (BYDV) 3'-untranslated region correlates with translation efficiency.
Banerjee B, Goss DJ. Banerjee B, et al. J Biol Chem. 2014 Feb 14;289(7):4286-94. doi: 10.1074/jbc.M113.530329. Epub 2013 Dec 30. J Biol Chem. 2014. PMID: 24379412 Free PMC article. - Cap-Independent Translation: What's in a Name?
Shatsky IN, Terenin IM, Smirnova VV, Andreev DE. Shatsky IN, et al. Trends Biochem Sci. 2018 Nov;43(11):882-895. doi: 10.1016/j.tibs.2018.04.011. Epub 2018 May 19. Trends Biochem Sci. 2018. PMID: 29789219 Review.
Cited by
- Ribosome Profiling: Global Views of Translation.
Ingolia NT, Hussmann JA, Weissman JS. Ingolia NT, et al. Cold Spring Harb Perspect Biol. 2019 May 1;11(5):a032698. doi: 10.1101/cshperspect.a032698. Cold Spring Harb Perspect Biol. 2019. PMID: 30037969 Free PMC article. Review. - Activation and execution of the hepatic integrated stress response by dietary essential amino acid deprivation is amino acid specific.
Jonsson WO, Mirek ET, Wek RC, Anthony TG. Jonsson WO, et al. FASEB J. 2022 Jul;36(7):e22396. doi: 10.1096/fj.202200204RR. FASEB J. 2022. PMID: 35690926 Free PMC article. - m6A reader proteins: the executive factors in modulating viral replication and host immune response.
Yang D, Zhao G, Zhang HM. Yang D, et al. Front Cell Infect Microbiol. 2023 May 30;13:1151069. doi: 10.3389/fcimb.2023.1151069. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37325513 Free PMC article. Review. - The emerging roles of N6-methyladenosine RNA methylation in human cancers.
Shen H, Lan Y, Zhao Y, Shi Y, Jin J, Xie W. Shen H, et al. Biomark Res. 2020 Jun 29;8:24. doi: 10.1186/s40364-020-00203-6. eCollection 2020. Biomark Res. 2020. PMID: 32612834 Free PMC article. Review. - A compact regulatory RNA element in mouse Hsp70 mRNA.
Wang W, Liu F, Ugalde MV, Pyle AM. Wang W, et al. NAR Mol Med. 2024 Jan 29;1(1):ugae002. doi: 10.1093/narmme/ugae002. eCollection 2024 Jan. NAR Mol Med. 2024. PMID: 38318492 Free PMC article.
References
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol. 2006;2:213–220. - PubMed
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous