Unique Directional Motility of Influenza C Virus Controlled by Its Filamentous Morphology and Short-Range Motions - PubMed (original) (raw)
Unique Directional Motility of Influenza C Virus Controlled by Its Filamentous Morphology and Short-Range Motions
Tatsuya Sakai et al. J Virol. 2018.
Abstract
Influenza virus motility is based on cooperation between two viral spike proteins, hemagglutinin (HA) and neuraminidase (NA), and is a major determinant of virus infectivity. To translocate a virus particle on the cell surface, HA molecules exchange viral receptors and NA molecules accelerate the receptor exchange of HA. This type of virus motility was recently identified in influenza A virus (IAV). To determine if other influenza virus types have a similar receptor exchange mechanism-driven motility, we investigated influenza C virus (ICV) motility on a receptor-fixed glass surface. This system excludes receptor mobility, which makes it more desirable than a cell surface for demonstrating virus motility by receptor exchange. Like IAV, ICV was observed to move across the receptor-fixed surface. However, in contrast to the random movement of IAV, a filamentous ICV strain, Ann Arbor/1/50 (AA), moved in a straight line, in a directed manner, and at a constant rate, whereas a spherical ICV strain, Taylor/1233/47 (Taylor), moved randomly, similar to IAV. The AA and Taylor viruses each moved with a combination of gradual (crawling) and rapid (gliding) motions, but the distances of crawling and gliding for the AA virus were shorter than those of the Taylor virus. Our findings indicate that like IAV, ICV also has a motility that is driven by the receptor exchange mechanism. However, compared with IAV movement, filamentous ICV movement is highly regulated in both direction and speed. Control of ICV movement is based on its specific motility employing short crawling and gliding motions as well as its own filamentous morphology.IMPORTANCE Influenza virus enters into a host cell for infection via cellular endocytosis. Human influenza virus infects epithelial cells of the respiratory tract, the surfaces of which are hidden by abundant cilia that are inactive in endocytosis. An open question is the manner by which the virus migrates to endocytosis-active domains. In analyzing individual virus behaviors through single-virus tracking, we identified a novel function of the hemagglutinin and esterase of influenza C virus (ICV) as the motility machinery. Hemagglutinin iteratively exchanges a viral receptor, causing virus movement. Esterase degrades the receptors along the trajectory traveled by the virus and prevents the virus from moving backward, causing directional movement. We propose that ICV has a unique motile machinery directionally controlled via hemagglutinin sensing the receptor density manipulated by esterase.
Keywords: hemagglutinin-esterase-fusion glycoprotein; human influenza; influenza C virus; virus motility.
Copyright © 2018 Sakai et al.
Figures
FIG 1
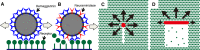
Hypotheses for influenza virus movement orientation mechanisms. (A) IAV moves on a cell surface by the HA-receptor exchange mechanism. (B) IAV NAs degrade receptor molecules along the pathway traveled by the virus, preventing the virus from moving backward and setting the direction of virus movement as forward. (C) A spherical IAV tends to move randomly. Receptor molecules are degraded by viral NAs in a narrow line along the virus trajectory. Because most receptors remain around the virus, the virus can move in all directions except for directly backward, resulting in a pattern of spherical virus movement that resembles random. (D) A filamentous ICV has the potential to move directionally. Receptors are two-dimensionally degraded by the ESs of ICV, which correspond to the NAs of IAV. The higher density of receptor molecules in front of the virus makes the virus move forward.
FIG 2
A novel imaging technique for analysis of virus motility. (A) Optics of SRICM. SRICM enhances the interference of light reflected by an interphase between the glass surface and virus particle and allows the imaging of unlabeled influenza viruses. (B) Representative SRICM images of unlabeled ICVs are shown. A filamentous ICV is observed as a rodlike object (arrow). Spherical ICVs are also seen (arrowheads). Bar, 1 μm. (C) Effects of anti-HA antibody on ICV movements. Three filamentous ICVs (black, gray, and white arrowheads) moved on the mucin-coated glass surface before the addition of anti-HA antibody. Representative images from various time points before (0 and 165 s) and after antibody addition (200 and 300 s) are shown. Time (in seconds) is indicated in the upper right of each panel. Bar, 1 μm. (D) Effects of anti-ES antibody on ICV movements. Two filamentous ICVs (black and white arrowheads) moved on the mucin-coated glass surface before the addition of anti-ES antibody. Representative images from various time points before (0 and 120 s) and after antibody addition (240 and 600 s) are shown. Time (in seconds) is indicated in the upper right of each panel. Bar, 1 μm.
FIG 3
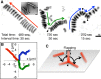
Filamentous ICV movements on a mucin-coated surface. (A) Superimposed images of filamentous ICVs as they move along a mucin-coated surface. Three different viruses are shown. Arrows indicate the directions of viral movements. The viruses either moved straight without turning (left), with occasional turns (middle), or with relatively frequent turns (right). Virus turns are indicated by arrowheads. Bar, 1 μm. (Inset) Electron microscopy (EM) images of negatively stained filamentous AA viruses. Arrowheads indicate bends in the virus filament. Bar, 0.1 μm. (B) Trajectories of the three ICVs shown in panel A. The centers of ICV were plotted every 1 s. The red, green, and blue trajectories correspond to the left, middle, and right ICVs, respectively, in panel A. (C) A schematic illustrating the movement of the filamentous ICV in the middle of panel A. While turning, the filamentous virus stood up, flapped for several seconds, and then flopped down. Thereafter, the virus moved at an angle different from that of its original direction of movement.
FIG 4
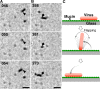
Spherical ICV movements on a mucin-coated surface. (A) Morphology of AA virus. Negatively stained AA viruses were observed by EM. Arrowheads and arrows indicate spherical and filamentous viruses, respectively. Bar, 0.5 μm. (Inset) High-magnification image of a single spherical AA virus. Arrowheads indicate some of the HEF spikes that are densely distributed over the entire virus surface. Bar, 0.1 μm. (B) Trajectories of three spherical AA viruses. (C) Enlarged image of the square in panel B. (D) EM images of Taylor viruses. Bar, 0.5 μm. (Inset) High-magnification image of a single spherical Taylor virus. Arrowheads indicate some of the HEF spikes that are densely distributed over the entire virus surface. Bar, 0.1 μm. (E) Trajectories of three spherical Taylor viruses. (F) Enlarged image of the square in panel E. Arrows indicate rapid (gliding) motions. In panels B, C, E, and F, the centers of the ICVs were plotted every 1 s.
FIG 5
Flapping of a short filamentous Taylor virus. (A, B) Two sequential images of the single filamentous virus in Movie S6 are shown. Arrows and arrowheads indicate the identical ends of the virus filament. For several seconds around 50 s (A) and 261 s (B), the filamentous virus was flapping. The time (in seconds) is indicated in the upper left of each panel. Bar, 1 μm. (C) A schematic illustrating virus flapping.
FIG 6
Quantitative analyses of ICV movements. (A) Mean square displacements (MSDs) of the filamentous AA, spherical AA, and spherical Taylor viruses. (B) Log-log plots of MSDs. MSDs were fitted by the equation: ⟨_R_2⟩ = C × tn, where ⟨_R_2⟩ is the MSD, C is the constant, t is time, and n is a constant between 1 and 2. Fitting curves are presented as blue or green lines. The lower graph is an enlarged graph of spherical AA virus MSD and corresponds to the square in the upper graph. (C) Frequency analyses of crawling and gliding by filamentous AA, spherical AA, and spherical Taylor viruses. The displacements of the viruses per second were analyzed using the complementary cumulative distribution functions (CCDF). Virus crawling and gliding phases are highlighted by blue and green, respectively. Lengths of crawling and gliding are expressed as means ± standard deviations (SD).
FIG 7
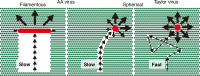
Dual regulation mechanisms of ICV directional movement. A spherical Taylor virus (red circle) moves quickly on the surface utilizing long crawling and gliding (right). During fast movement, viral ESs degrade only a small amount of receptor molecules (green circles) along the virus trajectory. After virus movement, most receptors remain around the virus and allow the virus to move in various directions, resulting in the random movement of the Taylor virus. In contrast, a spherical AA virus (red circle) moves slowly with short crawling and gliding, and viral ESs degrade a relatively large number of receptors along the trajectory (middle). Consequently, the virus tends to move forward. In a filamentous AA virus (red rod), the directional movement persists for a long period of time due to the filamentous morphology of the virus (left). Because the receptor molecules behind the virus filament are broadly degraded, the distinct difference in receptor density between the front and the back of the virus filament fixes the direction of virus movement.
Similar articles
- TMPRSS2 Activates Hemagglutinin-Esterase Glycoprotein of Influenza C Virus.
Sato K, Hayashi H, Shimotai Y, Yamaya M, Hongo S, Kawakami K, Matsuzaki Y, Nishimura H. Sato K, et al. J Virol. 2021 Oct 13;95(21):e0129621. doi: 10.1128/JVI.01296-21. Epub 2021 Aug 18. J Virol. 2021. PMID: 34406864 Free PMC article. - Influenza A virus hemagglutinin and neuraminidase act as novel motile machinery.
Sakai T, Nishimura SI, Naito T, Saito M. Sakai T, et al. Sci Rep. 2017 Mar 27;7:45043. doi: 10.1038/srep45043. Sci Rep. 2017. PMID: 28344335 Free PMC article. - Hemagglutinin-esterase-fusion (HEF) protein of influenza C virus.
Wang M, Veit M. Wang M, et al. Protein Cell. 2016 Jan;7(1):28-45. doi: 10.1007/s13238-015-0193-x. Epub 2015 Jul 28. Protein Cell. 2016. PMID: 26215728 Free PMC article. Review. - The Hemagglutinin-Esterase Fusion Glycoprotein Is a Primary Determinant of the Exceptional Thermal and Acid Stability of Influenza D Virus.
Yu J, Hika B, Liu R, Sheng Z, Hause BM, Li F, Wang D. Yu J, et al. mSphere. 2017 Aug 9;2(4):e00254-17. doi: 10.1128/mSphere.00254-17. eCollection 2017 Jul-Aug. mSphere. 2017. PMID: 28808690 Free PMC article. - Influenza A Virus Hemagglutinin-Neuraminidase-Receptor Balance: Preserving Virus Motility.
de Vries E, Du W, Guo H, de Haan CAM. de Vries E, et al. Trends Microbiol. 2020 Jan;28(1):57-67. doi: 10.1016/j.tim.2019.08.010. Epub 2019 Oct 17. Trends Microbiol. 2020. PMID: 31629602 Free PMC article. Review.
Cited by
- Hopping and crawling DNA-coated colloids.
Zheng JA, Holmes-Cerfon M, Pine DJ, Marbach S. Zheng JA, et al. Proc Natl Acad Sci U S A. 2024 Oct 8;121(41):e2318865121. doi: 10.1073/pnas.2318865121. Epub 2024 Oct 1. Proc Natl Acad Sci U S A. 2024. PMID: 39352927 - Unraveling dynamics of paramyxovirus-receptor interactions using nanoparticles displaying hemagglutinin-neuraminidase.
Wu X, Goebbels M, Debski-Antoniak O, Marougka K, Chao L, Smits T, Wennekes T, Kuppeveld FJMV, Vries E, de Haan CAM. Wu X, et al. PLoS Pathog. 2024 Jul 25;20(7):e1012371. doi: 10.1371/journal.ppat.1012371. eCollection 2024 Jul. PLoS Pathog. 2024. PMID: 39052678 Free PMC article. - Worm blobs as entangled living polymers: from topological active matter to flexible soft robot collectives.
Deblais A, Prathyusha KR, Sinaasappel R, Tuazon H, Tiwari I, Patil VP, Bhamla MS. Deblais A, et al. Soft Matter. 2023 Sep 27;19(37):7057-7069. doi: 10.1039/d3sm00542a. Soft Matter. 2023. PMID: 37706563 Free PMC article. Review. - Receptor Density-Dependent Motility of Influenza Virus Particles on Surface Gradients.
Hamming PHE, Overeem NJ, Diestelhorst K, Fiers T, Tieke M, Vos GM, Boons GPH, van der Vries E, Block S, Huskens J. Hamming PHE, et al. ACS Appl Mater Interfaces. 2023 May 24;15(20):25066-25076. doi: 10.1021/acsami.3c05299. Epub 2023 May 11. ACS Appl Mater Interfaces. 2023. PMID: 37167605 Free PMC article. - Kinetic analysis of paramyxovirus-sialoglycan receptor interactions reveals virion motility.
Wu X, Goebbels M, Chao L, Wennekes T, van Kuppeveld FJM, de Vries E, de Haan CAM. Wu X, et al. PLoS Pathog. 2023 Mar 27;19(3):e1011273. doi: 10.1371/journal.ppat.1011273. eCollection 2023 Mar. PLoS Pathog. 2023. PMID: 36972304 Free PMC article.
References
- Shaw ML, Palese P. 2013. Orthomyxoviridae, p 1151–1185. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA.
- Cox NJ, Neumann G, Donis RO, Kawaoka Y. 2010. Orthomyxoviruses: influenza, p 1–65. In Mahy BWJ, ter Meulen V, Borriello SP, Murray PR, Funke G, Merz WG, Hay RJ, Cox FEG, Despommier DD, Kaufmann SHE, Steward MW (ed), Topley and Wilson's microbiology and microbial infections; Wiley, Hoboken, NJ.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources