Critical Role of Plasmacytoid Dendritic Cells in Regulating Gene Expression and Innate Immune Responses to Human Rhinovirus-16 - PubMed (original) (raw)
Critical Role of Plasmacytoid Dendritic Cells in Regulating Gene Expression and Innate Immune Responses to Human Rhinovirus-16
Yang Xi et al. Front Immunol. 2017.
Abstract
Though human rhinoviruses (HRVs) are usually innocuous viruses, they can trigger serious consequences in certain individuals, especially in the setting of impaired interferon (IFN) synthesis. Plasmacytoid dendritic cells (pDCs) are key IFN producing cells, though we know little about the role of pDC in HRV-induced immune responses. Herein, we used gene expression microarrays to examine HRV-activated peripheral blood mononuclear cells (PBMCs) from healthy people, in combination with pDC depletion, to assess whether observed gene expression patterns were pDC dependent. As expected, pDC depletion led to a major reduction in IFN-α release. This was associated with profound differences in gene expression between intact PBMC and pDC-depleted PBMC, and major changes in upstream regulators: 70-80% of the HRV activated genes appeared to be pDC dependent. Real-time PCR confirmed key changes in gene expression, in which the following selected genes were shown to be highly pDC dependent: the transcription factor IRF7, both IL-27 chains (IL-27p28 and EBI3), the alpha chain of the IL-15 receptor (IL-15RA) and the IFN-related gene IFI27. HRV-induced IL-6, IFN-γ, and IL-27 protein synthesis were also highly pDC dependent. Supplementing pDC-depleted cultures with recombinant IL-15, IFN-γ, IL-27, or IL-6 was able to restore the IFN-α response, thereby compensating for the absence of pDC. Though pDC comprise only a minority population of migratory leukocytes, our findings highlight the profound extent to which these cells contribute to the immune response to HRV.
Keywords: human rhinovirus; human rhinovirus responsive genes; innate immune response; plasmacytoid dendritic cell-dependent gene expression; plasmacytoid dendritic cells.
Figures
Figure 1
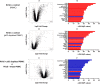
In response to human rhinoviruses strain 16 (RV16) stimulation, plasmacytoid dendritic cell (pDC)-depleted peripheral blood mononuclear cells (PBMCs) produce significantly less interferon (IFN)-α than intact PBMC. PBMC were either depleted of pDC using CD303 immunomagnetic beads or left intact. Efficiency of pDC depletion was examined with flow cytometry (A). PBMC (5 × 105 cells/well) were then cultured in the presence/absence of RV16 (MOI = 1) for 24 h at 37°C. IFN-α production in culture supernatant was measured by enzyme-linked immunosorbent assay (ELISA). Data shown are for the net RV16 simulated IFN-α concentration following subtraction of the IFN-α concentration in the absence of RV16 (media control). The dotted lines represent the median of each group (n = 15). IFN-α concentrations in pDC-depleted PBMC were significantly lower than intact PBMC (p < 0.001) (B).
Figure 2
In the presence of human rhinoviruses strain 16 (RV16), intact peripheral blood mononuclear cell (PBMC) and plasmacytoid dendritic cell (pDC)-depleted PBMCs exhibit distinct patterns of gene expression. Intact PBMC and pDC-depleted PBMC (n = 12) were cultured with/without RV16 for 24 h. Whole-genome transcriptional profiling was performed using Illumina Human HT-12 microarray. Hierarchical cluster analysis was employed to cluster genes and samples based on the similarity of their expression patterns. RV16 response patterns in PBMC and pDC-depleted PBMC from healthy individuals (n = 11) were analyzed by principal component analysis. RV16-stimulated PBMC are shown in red, unstimulated PBMC are black circles. RV16-stimulated pDC-depleted PBMC are shown in yellow, and unstimulated pDC-depleted PBMC are green circles (A). The heatmap illustrates the distinct difference in the RV16 induced gene expression in PBMC and pDC-depleted PBMC (B).
Figure 3
Molecular drivers of human rhinoviruses strain 16 (RV16) induced gene expression. Differentially expressed genes/molecular drivers were identified in peripheral blood mononuclear cell (PBMC) (A,D) and plasmacytoid dendritic cell (pDC)-depleted PBMC (B,E) in the presence of RV16. Comparison of RV16-stimulated pDC-depleted PBMC and intact PBMC (C,F). Data analysis was performed by limma (A–C) and upstream regulator analysis (D–F). The dashed horizontal line in (A–C) indicates FDR < 0.05. Drivers in red are predicted to be activated and those in blue are inhibited.
Figure 4
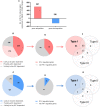
Which human rhinoviruses strain 16 (RV16) responsive genes are dependent on Plasmacytoid dendritic cell (pDC) and regulated by interferon (IFN)-I? RV16 responsive genes showing >2-fold changes have been analyzed (A). Out of the total genes that are upregulated (Red) or downregulated (Blue), the numbers of pDC-dependent gene expressions have been analyzed by comparing pDC-depleted PBMC with intact PBMC. Genes that show >2-fold change are regarded as “likely to be pDC dependent”; between 1.5- and 2.0-fold changes are regarded as “possibly pDC dependent”; and <1.5-fold change and regarded as “unlikely to be pDC dependent” Numbers of genes are indicated on the pie charts **(B,C)**. Out of the total genes that are likely to be pDC-dependent expression (>2-fold), the proportion of IFN-I-regulated gene expression has been identified using Interferome v2.01 database: (D,E). Venn diagrams show the extent to which IFN-I-regulated genes are regulated solely by IFN-I, or also regulated by multiple IFN subtypes (F,G).
Figure 5
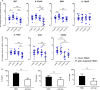
Validations experiments using real-time PCR (RT-PCR) and enzyme-linked immunosorbent assay (ELISA). Peripheral blood mononuclear cell (PBMC) and plasmacytoid dendritic cell (pDC)-depleted PBMC (n = 15) were cultured for 24 h in the absence or presence of human rhinoviruses strain 16 (RV16). mRNA expression of IRF7, IL-27p28, EBI3, IL-12p35, IL-15RA, IFI27, and CD303 was determined by RT-PCR. Data represent fold change relative to median of controls against the average of two reference genes (UBE2D2 and B2M) (A). IL-6 and interferon (IFN)-γ were measured in culture supernatant by ELISA. Net cytokine values are shown (RV16-stimulated cultures minus the unstimulated cultures) (B). All data represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 (I, intact PBMC; P, pDC-depleted PBMC).
Figure 6
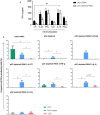
Addition of certain cytokines to plasmacytoid dendritic cell (pDC)-depleted cultures successfully rescues the interferon (IFN)-α response. Intact peripheral blood mononuclear cell (PBMC) (black bar) and pDC-depleted PBMC (white bar) were precultured with IL-27, IFN-γ, IL-6, IL-15, or IFN-β recombinant protein or with media only for 4 h at 37°C. All samples were subsequently stimulated with human rhinoviruses strain 16 (RV16) [multiplicity of infection (MOI) = 1] for a further 20 h. IFN-α protein was measured by ELISA (*p < 0.05; **p < 0.01; UT, untreated.) (A), and IFN-α producing cell subsets were measured by Flow Cytometry (*p < 0.05) (B). Data represent mean ± SD.
Similar articles
- Innate IFNs and plasmacytoid dendritic cells constrain Th2 cytokine responses to rhinovirus: a regulatory mechanism with relevance to asthma.
Pritchard AL, Carroll ML, Burel JG, White OJ, Phipps S, Upham JW. Pritchard AL, et al. J Immunol. 2012 Jun 15;188(12):5898-905. doi: 10.4049/jimmunol.1103507. Epub 2012 May 18. J Immunol. 2012. PMID: 22611238 - Eosinophil-mediated suppression and anti-IL-5 enhancement of plasmacytoid dendritic cell interferon responses in asthma.
Dill-McFarland KA, Schwartz JT, Zhao H, Shao B, Fulkerson PC, Altman MC, Gill MA. Dill-McFarland KA, et al. J Allergy Clin Immunol. 2022 Sep;150(3):666-675. doi: 10.1016/j.jaci.2022.03.025. Epub 2022 Apr 9. J Allergy Clin Immunol. 2022. PMID: 35413355 - Enhanced plasmacytoid dendritic cell antiviral responses after omalizumab.
Gill MA, Liu AH, Calatroni A, Krouse RZ, Shao B, Schiltz A, Gern JE, Togias A, Busse WW. Gill MA, et al. J Allergy Clin Immunol. 2018 May;141(5):1735-1743.e9. doi: 10.1016/j.jaci.2017.07.035. Epub 2017 Sep 1. J Allergy Clin Immunol. 2018. PMID: 28870461 Free PMC article. Clinical Trial. - Recent insights into the implications of metabolism in plasmacytoid dendritic cell innate functions: Potential ways to control these functions.
Saas P, Varin A, Perruche S, Ceroi A. Saas P, et al. F1000Res. 2017 Apr 10;6:456. doi: 10.12688/f1000research.11332.2. eCollection 2017. F1000Res. 2017. PMID: 28580131 Free PMC article. Review. - Anti-interferon alpha treatment in SLE.
Kirou KA, Gkrouzman E. Kirou KA, et al. Clin Immunol. 2013 Sep;148(3):303-12. doi: 10.1016/j.clim.2013.02.013. Epub 2013 Mar 1. Clin Immunol. 2013. PMID: 23566912 Review.
Cited by
- IRF7-Associated Immunophenotypes Have Dichotomous Responses to Virus/Allergen Coexposure and OM-85-Induced Reprogramming.
de Jong E, Lauzon-Joset JF, Leffler J, Serralha M, Larcombe AN, Christophersen CT, Holt PG, Strickland DH, Bosco A. de Jong E, et al. Front Immunol. 2021 Jul 22;12:699633. doi: 10.3389/fimmu.2021.699633. eCollection 2021. Front Immunol. 2021. PMID: 34367159 Free PMC article. - Identification of key genes and pathways in regulating immune‑induced diseases of dendritic cells by bioinformatic analysis.
Zheng Y, Zheng X, Li S, Zhang H, Liu M, Yang Q, Zhang M, Sun Y, Wu J, Yu B. Zheng Y, et al. Mol Med Rep. 2018 Jun;17(6):7585-7594. doi: 10.3892/mmr.2018.8834. Epub 2018 Mar 29. Mol Med Rep. 2018. PMID: 29620200 Free PMC article. - Whole transcriptome analysis of high and low IFN-α producers reveals differential response patterns following rhinovirus stimulation.
Murray LM, Thillaiyampalam G, Xi Y, Cristino AS, Upham JW. Murray LM, et al. Clin Transl Immunology. 2021 Nov 17;10(11):e1356. doi: 10.1002/cti2.1356. eCollection 2021. Clin Transl Immunology. 2021. PMID: 34868584 Free PMC article. - Recent advances in understanding rhinovirus immunity.
Makris S, Johnston S. Makris S, et al. F1000Res. 2018 Sep 24;7:F1000 Faculty Rev-1537. doi: 10.12688/f1000research.15337.1. eCollection 2018. F1000Res. 2018. PMID: 30345002 Free PMC article. Review. - Natural Killer Cells and Host Defense Against Human Rhinoviruses Is Partially Dependent on Type I IFN Signaling.
van der Heide SL, Xi Y, Upham JW. van der Heide SL, et al. Front Cell Infect Microbiol. 2020 Oct 21;10:510619. doi: 10.3389/fcimb.2020.510619. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33194777 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials