Identification of New Features from Known Bacterial Protective Vaccine Antigens Enhances Rational Vaccine Design - PubMed (original) (raw)
Identification of New Features from Known Bacterial Protective Vaccine Antigens Enhances Rational Vaccine Design
Edison Ong et al. Front Immunol. 2017.
Abstract
With many protective vaccine antigens reported in the literature and verified experimentally, how to use the knowledge mined from these antigens to support rational vaccine design and study underlying design mechanism remains unclear. In order to address the problem, a systematic bioinformatics analysis was performed on 291 Gram-positive and Gram-negative bacterial protective antigens with experimental evidence manually curated in the Protegen database. The bioinformatics analyses evaluated included subcellular localization, adhesin probability, peptide signaling, transmembrane α-helix and β-barrel, conserved domain, Clusters of Orthologous Groups, and Gene Ontology functional annotations. Here we showed the critical role of adhesins, along with subcellular localization, peptide signaling, in predicting secreted extracellular or surface-exposed protective antigens, with mechanistic explanations supported by functional analysis. We also found a significant negative correlation of transmembrane α-helix to antigen protectiveness in Gram-positive and Gram-negative pathogens, while a positive correlation of transmembrane β-barrel was observed in Gram-negative pathogens. The commonly less-focused cytoplasmic and cytoplasmic membrane proteins could be potentially predicted with the help of other selection criteria such as adhesin probability and functional analysis. The significant findings in this study can support rational vaccine design and enhance our understanding of vaccine design mechanisms.
Keywords: adhesin probability; conserved domains; functional analysis; protective antigen; reverse vaccinology; subcellular localization; transmembrane proteins; vaccine design.
Figures
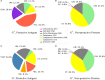
Figure 1
Subcellular localization profiles. G+ and G− bacterial protective antigens showed significantly higher (p < 0.01) proportions of EC (G+ and G−), CW (G+), and PE and OM (G−) (A,C) compared with the non-protective proteins (B,D) (abbreviations shown in the middle color key legend). Only PAgs and non-protective proteins are displayed and the composition of background proteomes and non-protective proteins are very alike (Figure S1 in Supplementary Material). The significant over-representation of PAgs’ subcellular localization prediction compared with non-protective proteins is indicated with “*” (p < 0.01).
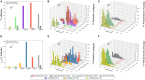
Figure 2
Profiles of adhesin probabilities of protective antigens (PAgs) and non-protective proteins with different subcellular localizations. The top three panels (A–C) show G+ pathogens, and the bottom three panels show G− pathogens. Specifically, the first column (A,D) represents the overall percentages of adhesin probabilities. The second column (B,E) and third column (C,F) show adhesin probability distributions of PAg and non-protective proteins, respectively. The red line in (B,C,E,F) indicates adhesin probability cutoff of no <0.51. Overall, PAgs have significantly higher (p < 0.01) percentages in EC (G+ and G−), CW (G+), and PE and OM (G−) (abbreviations shown in bottom color key legend). Interestingly, CM in G+ is also significant (p < 0.05) when coupled with adhesin probability. See the text for detailed discussion. The significant over-representation of PAgs’ adhesin probabilities at different subcellular localizations compared with non-protective proteins is indicated with “*” (p < 0.05) or “**” (p < 0.01).
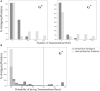
Figure 3
Transmembrane α-helix (G+ and G−) and β-barrel (G− only) profiles. As compared with non-protective proteins, there were much higher percentages of protective antigens with zero or one transmembrane α-helix (A). For transmembrane β-barrel (B), only two (0.0004%) out of all non-protective proteins had probability higher than the designated cutoff (indicated as black vertical line) described in method.
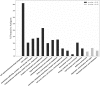
Figure 4
Over-representation of COG clustering profiles among reported protective antigens (PAgs). Over 40% of the reported PAgs belong to the cluster “Cell wall/membrane/envelop biogenesis,” which agrees with common knowledge of using surface-exposed proteins as a key criterion in vaccine antigen prediction. Other COG clusters related to pathogen motility, secretion, signal transduction, and transportation are also significantly enriched in PAgs as compared with non-protective proteins. See the text for detailed discussion. The significant over-representation of PAgs’ COG clusters compared with non-protective proteins is colored with gray (p < 0.05) and black (p < 0.01).
Figure 5
Over-representation of GO BP, MF, and CC term profiles among reported protective antigens (PAgs) and visualized using the GOfox tool. The number next to each GO term indicates the number of PAg with the corresponding GO functional annotation. Similar to COG clustering, GO terms that are related to pathogen motility, secretion, signal transduction, and transportation are also significantly enriched in PAgs as compared with non-protective proteins. The GO CC terms also supported the high preference of extracellular, surface-exposed (cell wall in G+ and outer membrane in G−), and periplasmic (G−) PAgs. The significant over-representation of PAgs’ GO terms compared with non-protective proteins is color-coded following the legend in the lower right corner.
Similar articles
- COVID-19 vaccine design using reverse and structural vaccinology, ontology-based literature mining and machine learning.
Huffman A, Ong E, Hur J, D'Mello A, Tettelin H, He Y. Huffman A, et al. Brief Bioinform. 2022 Jul 18;23(4):bbac190. doi: 10.1093/bib/bbac190. Brief Bioinform. 2022. PMID: 35649389 Free PMC article. - Antibiotic Resistance Determinant-Focused Acinetobacter baumannii Vaccine Designed Using Reverse Vaccinology.
Ni Z, Chen Y, Ong E, He Y. Ni Z, et al. Int J Mol Sci. 2017 Feb 21;18(2):458. doi: 10.3390/ijms18020458. Int J Mol Sci. 2017. PMID: 28230771 Free PMC article. - Systematic collection, annotation, and pattern analysis of viral vaccines in the VIOLIN vaccine knowledgebase.
Huffman A, Gautam M, Gandhi A, Du P, Austin L, Roan K, Zheng J, He Y. Huffman A, et al. Front Cell Infect Microbiol. 2025 Feb 7;15:1509226. doi: 10.3389/fcimb.2025.1509226. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 39991713 Free PMC article. - Surface and Exoproteomes of Gram-Positive Pathogens for Vaccine Discovery.
Biagini M, Bagnoli F, Norais N. Biagini M, et al. Curr Top Microbiol Immunol. 2017;404:309-337. doi: 10.1007/82_2016_50. Curr Top Microbiol Immunol. 2017. PMID: 28204975 Review. - Neutralizing antibody and functional mapping of Bacillus anthracis protective antigen-The first step toward a rationally designed anthrax vaccine.
McComb RC, Martchenko M. McComb RC, et al. Vaccine. 2016 Jan 2;34(1):13-9. doi: 10.1016/j.vaccine.2015.11.025. Epub 2015 Nov 21. Vaccine. 2016. PMID: 26611201 Review.
Cited by
- COVID-19 vaccine design using reverse and structural vaccinology, ontology-based literature mining and machine learning.
Huffman A, Ong E, Hur J, D'Mello A, Tettelin H, He Y. Huffman A, et al. Brief Bioinform. 2022 Jul 18;23(4):bbac190. doi: 10.1093/bib/bbac190. Brief Bioinform. 2022. PMID: 35649389 Free PMC article. - COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning.
Ong E, Wong MU, Huffman A, He Y. Ong E, et al. bioRxiv [Preprint]. 2020 Mar 22:2020.03.20.000141. doi: 10.1101/2020.03.20.000141. bioRxiv. 2020. PMID: 32511333 Free PMC article. Updated. Preprint. - Comparative Reverse Vaccinology of Piscirickettsia salmonis, Aeromonas salmonicida, Yersinia ruckeri, Vibrio anguillarum and Moritella viscosa, Frequent Pathogens of Atlantic Salmon and Lumpfish Aquaculture.
Chukwu-Osazuwa J, Cao T, Vasquez I, Gnanagobal H, Hossain A, Machimbirike VI, Santander J. Chukwu-Osazuwa J, et al. Vaccines (Basel). 2022 Mar 18;10(3):473. doi: 10.3390/vaccines10030473. Vaccines (Basel). 2022. PMID: 35335104 Free PMC article. - Cwp22, a novel peptidoglycan cross-linking enzyme, plays pleiotropic roles in Clostridioides difficile.
Zhu D, Bullock J, He Y, Sun X. Zhu D, et al. Environ Microbiol. 2019 Aug;21(8):3076-3090. doi: 10.1111/1462-2920.14706. Epub 2019 Jun 28. Environ Microbiol. 2019. PMID: 31173438 Free PMC article. - Emerging Concepts and Technologies in Vaccine Development.
Brisse M, Vrba SM, Kirk N, Liang Y, Ly H. Brisse M, et al. Front Immunol. 2020 Sep 30;11:583077. doi: 10.3389/fimmu.2020.583077. eCollection 2020. Front Immunol. 2020. PMID: 33101309 Free PMC article. Review.
References
- WHO. MDG 6: Combat HIV/AIDS, Malaria and Other Diseases. Geneva: WHO; (2014).
LinkOut - more resources
Full Text Sources
Other Literature Sources