Transcriptional integration of mitogenic and mechanical signals by Myc and YAP - PubMed (original) (raw)
. 2017 Oct 15;31(20):2017-2022.
doi: 10.1101/gad.301184.117. Epub 2017 Nov 15.
Serena De Fazio 1, Francesca Biagioni 1, Elisa Donato 1, Marieta Caganova 1, Laura Curti 1, Mirko Doni 2, Silvia Sberna 1, Deborah Aldeghi 1, Chiara Biancotto 1, Alessandro Verrecchia 2, Daniela Olivero 3, Bruno Amati 1 2, Stefano Campaner 1
Affiliations
- PMID: 29141911
- PMCID: PMC5733494
- DOI: 10.1101/gad.301184.117
Transcriptional integration of mitogenic and mechanical signals by Myc and YAP
Ottavio Croci et al. Genes Dev. 2017.
Abstract
Mammalian cells must integrate environmental cues to determine coherent physiological responses. The transcription factors Myc and YAP-TEAD act downstream from mitogenic signals, with the latter responding also to mechanical cues. Here, we show that these factors coordinately regulate genes required for cell proliferation. Activation of Myc led to extensive association with its genomic targets, most of which were prebound by TEAD. At these loci, recruitment of YAP was Myc-dependent and led to full transcriptional activation. This cooperation was critical for cell cycle entry, organ growth, and tumorigenesis. Thus, Myc and YAP-TEAD integrate mitogenic and mechanical cues at the transcriptional level to provide multifactorial control of cell proliferation.
Keywords: Hippo signaling; Myc; TEAD; YAP; transcription.
© 2017 Croci et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
Figure 1.
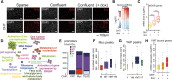
Myc and YAP coregulate cell cycle entry. Serum-starved subconfluent (sparse) (A) or highly confluent (confluent) 3T9MycER;YAP (B_–_H) cells were treated with OHT to activate MycER and doxycycline (dox) to trigger the expression of YAPS127A. (A) Cell cycle entry was measured by immunofluorescence analysis of EdU incorporation. DAPI was used to color nuclei. (B) Ranked heat map based on the log2 fold change of the differentially expressed genes (DEGs) identified by RNA sequencing (RNA-seq). (C) Box plot of the mRNA expression level of the Myc-dependent serum response (MDSR) genes (D) Gene ontology map based on the DEGs determined upon both MycER activation and YAP induction. (E) Cumulative bar graph of Myc, YAP, and TEAD ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) peaks, color-coded based on their overlap. (Y) YAP; (M) Myc; (T) TEAD. (F,G) Box plot of the enrichment of Myc (F) and YAP (G) ChIP-seq peaks divided into subsets as in E. (H) Expression levels of up-regulated genes cobound at their promoters by YAP, Myc, and TEAD (YMT peaks).
Figure 2.
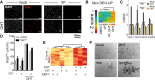
Myc-driven cell cycle entry depends on YAP activity and cytoskeletal tension. (A_–_E) Serum-starved subconfluent fibroblasts were kept in low serum and treated as indicated. (A) Immunofluorescence analysis of Myc-induced cell cycle entry of 3T9MycER measured as EdU incorporation on cells treated with the YAP inhibitor verteporfin (VP). (B) Expression analysis (clustering) of Myc up-regulated genes following VP treatment. (C) RT-qPCR expression of Myc target genes in MycER fibroblasts either wild type (YAP+/+) or knockout (Yap−/−) for Yap. (D) S-phase entry by BrdU incorporation (by FACS) in MycER fibroblasts overexpressing YAPS127A/S318A. Cells were treated with OHT to activate MycER and with the ROCK inhibitor Y276632 (Y27) as indicated. (E) Clustered heat map of normalized mRNA expression of cells shown in D. (F) Anchorage-independent growth assay of bipotential mouse embryonic liver (BMEL) cells overexpressing MycER and tet-YAPS127A, treated as indicated. Representative pictures of cell colonies are shown.
Figure 3.
Cooperative binding and transcriptional activation by Myc, TEAD, and YAP. Genome-wide analyses of livers from R26-rtTA mice either wild type (wt), transgenics for Myc (tet-Myc) or YAP (tet-YAP), or double transgenics (tet-Myc/YAP). Short-term induction was achieved by feeding mice with doxycycline-containing food for 48 h. (A) Liver sections stained with an anti-Ki67 antibody. (B) Hierarchical clustering of DEGs. (C) Box plot showing a representative cluster of YAP/Myc DEGs. (D) Venn analysis of Myc, YAP, and TEAD ChIP-seq peaks. The number of peaks determined for each TF is reported in brackets; the arrows point to the number of overlapping peaks. (E) Ranked heat maps of the ChIP-seq enrichment of the indicated TFs. (Top panel) YAP peaks detected only in tet-YAP livers. (Bottom panel) Promoters bound by YAP in tet-Myc/YAP livers. (F,G) H3K27ac (F) and H3K4me3 (G) levels at promoters of DEG-up genes cobound by Myc and YAP.
Figure 4.
Myc and YAP cooperate in inducing liver growth and tumorigenesis. (A) Liver weight assessed at 5 wk of induction. Data are reported as percentage relative to total body weight. (B) Kaplan-Meier disease-free survival analysis. (C) Western blotting analysis of YAP and Myc levels in LAP-tTA tet-YAP mice at the pretumoral stage (4 wk of YAP activation) and in tumors. Vinculin (vin) was used as aninternal control for equal loading. (D) Box plot of the expression level of MDSR genes up-regulated in the liver upon YAP and/or Myc induction. (Inset at the right) Ranked heat map. (E, top panel) Heat map of Myc and YAP/TAZ gene signatures based on the expression data of breast cancers (TCGA_BRCA). The heat map was clustered by breast cancer subtypes (basal-like, normal-like, and Luminal/Her2+). (Bottom panel) The statistical track shows the logarithmic plot of _P_-values for each gene. (Red bars) Genes up in basal-like; (green bars) genes up in Luminal/Her+.
Similar articles
- Mutual dependence of the MRTF-SRF and YAP-TEAD pathways in cancer-associated fibroblasts is indirect and mediated by cytoskeletal dynamics.
Foster CT, Gualdrini F, Treisman R. Foster CT, et al. Genes Dev. 2017 Dec 1;31(23-24):2361-2375. doi: 10.1101/gad.304501.117. Epub 2018 Jan 9. Genes Dev. 2017. PMID: 29317486 Free PMC article. - Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling.
Ota M, Sasaki H. Ota M, et al. Development. 2008 Dec;135(24):4059-69. doi: 10.1242/dev.027151. Epub 2008 Nov 12. Development. 2008. PMID: 19004856 - Cell competition in mouse NIH3T3 embryonic fibroblasts is controlled by the activity of Tead family proteins and Myc.
Mamada H, Sato T, Ota M, Sasaki H. Mamada H, et al. J Cell Sci. 2015 Feb 15;128(4):790-803. doi: 10.1242/jcs.163675. Epub 2015 Jan 14. J Cell Sci. 2015. PMID: 25588835 - YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth.
Low BC, Pan CQ, Shivashankar GV, Bershadsky A, Sudol M, Sheetz M. Low BC, et al. FEBS Lett. 2014 Aug 19;588(16):2663-70. doi: 10.1016/j.febslet.2014.04.012. Epub 2014 Apr 18. FEBS Lett. 2014. PMID: 24747426 Review. - YAP is essential for 3D organogenesis withstanding gravity.
Asaoka Y, Nishina H, Furutani-Seiki M. Asaoka Y, et al. Dev Growth Differ. 2017 Jan;59(1):52-58. doi: 10.1111/dgd.12338. Epub 2017 Jan 16. Dev Growth Differ. 2017. PMID: 28093734 Review.
Cited by
- The oncogenic axis YAP/MYC/EZH2 impairs PTEN tumor suppression activity enhancing lung tumorigenicity.
Lo Sardo F, Turco C, Messina B, Sacconi A, Auciello FR, Pulito C, Strano S, Lev S, Blandino G. Lo Sardo F, et al. Cell Death Discov. 2024 Oct 25;10(1):452. doi: 10.1038/s41420-024-02216-8. Cell Death Discov. 2024. PMID: 39455556 Free PMC article. - Lymphocyte-Specific Protein-1 Suppresses Xenobiotic-Induced Constitutive Androstane Receptor and Subsequent Yes-Associated Protein-Activated Hepatocyte Proliferation.
Koral K, Bhushan B, Orr A, Stoops J, Bowen WC, Copeland MA, Locker J, Mars WM, Michalopoulos GK. Koral K, et al. Am J Pathol. 2022 Jun;192(6):887-903. doi: 10.1016/j.ajpath.2022.03.010. Epub 2022 Apr 4. Am J Pathol. 2022. PMID: 35390317 Free PMC article. - Caspase-1 and the inflammasome promote polycystic kidney disease progression.
Swenson-Fields KI, Ward CJ, Lopez ME, Fross S, Heimes Dillon AL, Meisenheimer JD, Rabbani AJ, Wedlock E, Basu MK, Jansson KP, Rowe PS, Stubbs JR, Wallace DP, Vitek MP, Fields TA. Swenson-Fields KI, et al. Front Mol Biosci. 2022 Nov 29;9:971219. doi: 10.3389/fmolb.2022.971219. eCollection 2022. Front Mol Biosci. 2022. PMID: 36523654 Free PMC article. - Transformation of normal cells by aberrant activation of YAP via cMyc with TEAD.
Nishimoto M, Uranishi K, Asaka MN, Suzuki A, Mizuno Y, Hirasaki M, Okuda A. Nishimoto M, et al. Sci Rep. 2019 Jul 29;9(1):10933. doi: 10.1038/s41598-019-47301-6. Sci Rep. 2019. PMID: 31358774 Free PMC article. - dNTP metabolism links mechanical cues and YAP/TAZ to cell growth and oncogene-induced senescence.
Santinon G, Brian I, Pocaterra A, Romani P, Franzolin E, Rampazzo C, Bicciato S, Dupont S. Santinon G, et al. EMBO J. 2018 Jun 1;37(11):e97780. doi: 10.15252/embj.201797780. Epub 2018 Apr 12. EMBO J. 2018. PMID: 29650681 Free PMC article.
References
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. 2013. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154: 1047–1059. - PubMed
- Armelin HA, Armelin MC, Kelly K, Stewart T, Leder P, Cochran BH, Stiles CD. 1984. Functional role for c-myc in mitogenic response to platelet-derived growth factor. Nature 310: 655–660. - PubMed
- Barone MV, Courtneidge SA. 1995. Myc but not Fos rescue of PDGF signalling block caused by kinase-inactive Src. Nature 378: 509–512. - PubMed
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. 2007. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol 17: 2054–2060. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases