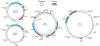Viruses of archaea: Structural, functional, environmental and evolutionary genomics - PubMed (original) (raw)
Review
Viruses of archaea: Structural, functional, environmental and evolutionary genomics
Mart Krupovic et al. Virus Res. 2018.
Abstract
Viruses of archaea represent one of the most enigmatic parts of the virosphere. Most of the characterized archaeal viruses infect extremophilic hosts and display remarkable diversity of virion morphotypes, many of which have never been observed among viruses of bacteria or eukaryotes. The uniqueness of the virion morphologies is matched by the distinctiveness of the genomes of these viruses, with ∼75% of genes encoding unique proteins, refractory to functional annotation based on sequence analyses. In this review, we summarize the state-of-the-art knowledge on various aspects of archaeal virus genomics. First, we outline how structural and functional genomics efforts provided valuable insights into the functions of viral proteins and revealed intricate details of the archaeal virus-host interactions. We then highlight recent metagenomics studies, which provided a glimpse at the diversity of uncultivated viruses associated with the ubiquitous archaea in the oceans, including Thaumarchaeota, Marine Group II Euryarchaeota, and others. These findings, combined with the recent discovery that archaeal viruses mediate a rapid turnover of thaumarchaea in the deep sea ecosystems, illuminate the prominent role of these viruses in the biosphere. Finally, we discuss the origins and evolution of archaeal viruses and emphasize the evolutionary relationships between viruses and non-viral mobile genetic elements. Further exploration of the archaeal virus diversity as well as functional studies on diverse virus-host systems are bound to uncover novel, unexpected facets of the archaeal virome.
Copyright © 2017 Elsevier B.V. All rights reserved.
Figures
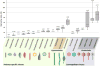
Figure 1
Genome size distribution and morphotypes of archaeal viruses. In the box plot, each box represents the middle 50th percentile of the data set and is derived using the lower and upper quartile values. The median value is displayed by a horizontal line inside the box. Whiskers represent the maximum and minimum values. The number of genomes used for construction of the box plot is indicated for each group of archaeal viruses. The information on the genome sizes was collected from the GenBank records as well as from the published literature (for genomes assembled from metagenomic data). Archaea-specific and cosmopolitan fractions of the archaeal virosphere are indicated with different background colors.
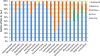
Figure 2
Fraction of archaeal virus proteins with best-hits to proteins encoded by other viruses or cellular organisms. BLASTP searches were performed using two inclusion thresholds, E<1e-05 and E<1e-03 (Table S1). The plot shown in the figure is created using the former E value cut-off. The classification of the hits is based on the best-hit only. Self-hits were eliminated for the calculations. Hits to the Sulfolobus acidocaldarius genome, which contains a provirus closely related to turriviruses STIV and STIV2, (Anderson et al., 2017) were excluded as well. The “Unclassified” category includes Sulfolobales viruses YNP1 and YNP2 (Gudbergsdóttir et al., 2016), ANMV-1 (Paul et al., 2015), Hyperthermophilic archaeal virus 1 (Garrett et al., 2010), Pyrococcus abyssi virus 1 (Geslin et al., 2007) and Thermococcus prieurii virus 1 (Gorlas et al., 2012).
Figure 3
Genome comparison of pleolipoviruses and salterproviruses. The genome schematics are drawn roughly to scale (shown at the bottom of the figure). The genes are shown by block arrows indicating the direction of transcription. The genes encoding for genome replication-associated proteins and virion proteins color coded: structural proteins of pleolipoviruses and salterproviruses are shown in light and dark green, respectively; two families of rolling circle replication initiation endonucleases (RCRE1 and RCRE2) are colored red and magenta, respectively; uncharacterized Rep is shown in orange; protein-primed family B DNA polymerase (pPolB), light blue. Genes shared between viruses are indicated by yellow shading. Morphologies of the corresponding viruses are shown on the right of the figure. MCP, major capsid protein. Genome accession numbers: HRPV-1, FJ685651; HHPV-1, GU321093; HRPV-3, JN882265; His2, AF191797; His1, AF191796; His1-like contig 5357, LFUF01004316 (BioProject PRJNA287316).
Figure 4
Comparative genomics of archaeal viruses, represented as a network of genomes and shared genes. A. The gene sharing network of the entire dsDNA virosphere highlights the distinct nature of the archaeal-specific viruses. Each viral genome is represented as a circle (colored circles correspond to archaeal viruses). Straight lines (edges) connect each virus with the gene families present in its genome. Consequently, edge junctions denote the presence of a gene family shared by multiple viruses. In this bipartite representation, closely related genomes are connected indirectly through a large number of shared gene families. The network has been projected onto a plane to show groups of similar genomes close to each other. Different shades of grey represent the four supermodules of the dsDNA virosphere, from lighter to darker: (i) the double-jelly roll fold MCP supermodule (including “Megavirales”, Adenoviridae and polintons, among others), (ii) the HK97-like MCP supermodule (including Caudovirales and Herpesvirales), (iii) Papillomaviruses and Polyomaviruses, and (iv) Baculo-like viruses. The color scheme for archaeal genomes follows the classification in modules shown in panel B. The high density of edges in the main body of the network results from the widespread gene sharing among most bacterial and eukaryotic viral groups, as well as the subset of archaeal viruses that are related to tailed bacteriophages (Caudovirales). In contrast, the archaeal-specific portion of the network is characterized by the presence of largely isolated clusters of similar genomes (modules) that correspond to distinct taxa. The figure is modified from (Iranzo et al., 2016b). B. Modular structure and gene sharing patterns among archaeal viruses. Groups of closely related genomes (modules) are highlighted with different colors. Black edges indicate the connections that involve broadly shared gene families (connector genes). Within each module, colored edges indicate the presence of “signature” genes, i.e. genes that are diagnostic of the members of the given module. The network also includes some capsid-less mobile genetic elements related to archaeal viruses and discussed in the text, such as casposons, which are represented as triangles. GT, glycosyltransferase of the GT-B superfamily; RHH, ribbon-helix-helix domain-containing protein; pPolB, protein-primed DNA polymerase B; MCP, major capsid protein; HAV1, Hyperthermophilic archaeal virus 1. The figure is updated from (Iranzo et al., 2016a). The original data set was supplemented with 26 genomes of Magroviruses (Philosof et al., 2017), 3 genomes of uncultivated His1-like viruses (Adriaenssens et al., 2016), 1 genome of an uncultivated member of the Caudovirales from the Oceanic basement (Nigro et al., 2017), and 1 representative genome of the family Portogloboviridae (Liu et al., 2017).
Figure 5
Relationship between thermococcal plasmids and viruses. Homologous genes are colored similarly. PAV1 ORF528, which has homologues in haloarchaeal plasmids, is shaded grey. Note that viruses PAV1 and TPV1 share only genes encoding structural proteins (dark green arrows). Abbreviations, MCP, major capsid protein; SF1, superfamily 1; wHTH, winged helix-turn-helix; RHH, ribbon-helix-helix; HJR, Holliday junction resolvase; prim-pol, primase-polymerase.
Similar articles
- Structure and assembly of archaeal viruses.
Baquero DP, Liu Y, Wang F, Egelman EH, Prangishvili D, Krupovic M. Baquero DP, et al. Adv Virus Res. 2020;108:127-164. doi: 10.1016/bs.aivir.2020.09.004. Epub 2020 Oct 5. Adv Virus Res. 2020. PMID: 33837715 Review. - Bipartite Network Analysis of the Archaeal Virosphere: Evolutionary Connections between Viruses and Capsidless Mobile Elements.
Iranzo J, Koonin EV, Prangishvili D, Krupovic M. Iranzo J, et al. J Virol. 2016 Nov 28;90(24):11043-11055. doi: 10.1128/JVI.01622-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27681128 Free PMC article. - Archaeal viruses: living fossils of the ancient virosphere?
Prangishvili D. Prangishvili D. Ann N Y Acad Sci. 2015 Apr;1341:35-40. doi: 10.1111/nyas.12710. Epub 2015 Feb 25. Ann N Y Acad Sci. 2015. PMID: 25716458 - The enigmatic archaeal virosphere.
Prangishvili D, Bamford DH, Forterre P, Iranzo J, Koonin EV, Krupovic M. Prangishvili D, et al. Nat Rev Microbiol. 2017 Nov 10;15(12):724-739. doi: 10.1038/nrmicro.2017.125. Nat Rev Microbiol. 2017. PMID: 29123227 Review. - Postcards from the edge: structural genomics of archaeal viruses.
Krupovic M, White MF, Forterre P, Prangishvili D. Krupovic M, et al. Adv Virus Res. 2012;82:33-62. doi: 10.1016/B978-0-12-394621-8.00012-1. Adv Virus Res. 2012. PMID: 22420850 Review.
Cited by
- New archaeal viruses discovered by metagenomic analysis of viral communities in enrichment cultures.
Liu Y, Brandt D, Ishino S, Ishino Y, Koonin EV, Kalinowski J, Krupovic M, Prangishvili D. Liu Y, et al. Environ Microbiol. 2019 Jun;21(6):2002-2014. doi: 10.1111/1462-2920.14479. Epub 2019 Jan 8. Environ Microbiol. 2019. PMID: 30451355 Free PMC article. - Metagenomic analysis reveals unexplored diversity of archaeal virome in the human gut.
Li R, Wang Y, Hu H, Tan Y, Ma Y. Li R, et al. Nat Commun. 2022 Dec 29;13(1):7978. doi: 10.1038/s41467-022-35735-y. Nat Commun. 2022. PMID: 36581612 Free PMC article. - Integrated mobile genetic elements in Thaumarchaeota.
Krupovic M, Makarova KS, Wolf YI, Medvedeva S, Prangishvili D, Forterre P, Koonin EV. Krupovic M, et al. Environ Microbiol. 2019 Jun;21(6):2056-2078. doi: 10.1111/1462-2920.14564. Epub 2019 Mar 18. Environ Microbiol. 2019. PMID: 30773816 Free PMC article. - A new type of DNA phosphorothioation-based antiviral system in archaea.
Xiong L, Liu S, Chen S, Xiao Y, Zhu B, Gao Y, Zhang Y, Chen B, Luo J, Deng Z, Chen X, Wang L, Chen S. Xiong L, et al. Nat Commun. 2019 Apr 11;10(1):1688. doi: 10.1038/s41467-019-09390-9. Nat Commun. 2019. PMID: 30975999 Free PMC article.
References
- Abrescia NG, Bamford DH, Grimes JM, Stuart DI. Structure unifies the viral universe. Annu Rev Biochem. 2012;81:795–822. - PubMed
- Ahn DG, Kim SI, Rhee JK, Kim KP, Pan JG, Oh JW. TTSV1, a new virus-like particle isolated from the hyperthermophilic crenarchaeote Thermoproteus tenax. Virology. 2006;351(2):280–90. - PubMed
- Arnold HP, Ziese U, Zillig W. SNDV, a novel virus of the extremely thermophilic and acidophilic archaeon Sulfolobus. Virology. 2000;272(2):409–16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources