Global proteomics profiling improves drug sensitivity prediction: results from a multi-omics, pan-cancer modeling approach - PubMed (original) (raw)
Global proteomics profiling improves drug sensitivity prediction: results from a multi-omics, pan-cancer modeling approach
Mehreen Ali et al. Bioinformatics. 2018.
Abstract
Motivation: Proteomics profiling is increasingly being used for molecular stratification of cancer patients and cell-line panels. However, systematic assessment of the predictive power of large-scale proteomic technologies across various drug classes and cancer types is currently lacking. To that end, we carried out the first pan-cancer, multi-omics comparative analysis of the relative performance of two proteomic technologies, targeted reverse phase protein array (RPPA) and global mass spectrometry (MS), in terms of their accuracy for predicting the sensitivity of cancer cells to both cytotoxic chemotherapeutics and molecularly targeted anticancer compounds.
Results: Our results in two cell-line panels demonstrate how MS profiling improves drug response predictions beyond that of the RPPA or the other omics profiles when used alone. However, frequent missing MS data values complicate its use in predictive modeling and required additional filtering, such as focusing on completely measured or known oncoproteins, to obtain maximal predictive performance. Rather strikingly, the two proteomics profiles provided complementary predictive signal both for the cytotoxic and targeted compounds. Further, information about the cellular-abundance of primary target proteins was found critical for predicting the response of targeted compounds, although the non-target features also contributed significantly to the predictive power. The clinical relevance of the selected protein markers was confirmed in cancer patient data. These results provide novel insights into the relative performance and optimal use of the widely applied proteomic technologies, MS and RPPA, which should prove useful in translational applications, such as defining the best combination of omics technologies and marker panels for understanding and predicting drug sensitivities in cancer patients.
Availability and implementation: Processed datasets, R as well as Matlab implementations of the methods are available at https://github.com/mehr-een/bemkl-rbps.
Contact: mehreen.ali@helsinki.fi or tero.aittokallio@fimm.fi.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
Fig. 1.
Data modeling approach, employing BEMKL method, applied on NCI60 genomics (point mutations and CNV), molecular (gene and miRNA expression) and proteomics (MS and RPPA) profiles across 58 pan-cancer cell lines to predict drug response of selected drugs. The BEMKL method learns the multi-view kernel weights to form a joint kernelized representation of the data which is used with the multi-task drug weights to model the response profile across the cell lines to each individual drug (the outcome matrix at the right)
Fig. 2.
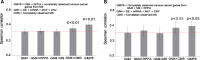
Average Spearman correlation, with standard error of the mean, between experimental and predicted drug sensitivity levels over 58 pan-cancer NCI-60 cell lines, using different omics data combinations for selected set of (A) 47 cytotoxic and (B) 24 targeted compounds. The red horizontal line indicates the baseline GM4 prediction accuracy. Statistical significance of the difference against the GM4 predictions was assessed with one-sided, paired _t_-test for the cytotoxic drugs and Wilcoxon signed-rank test for the targeted compounds. Statistical testing method was chosen based on the normality of the drug response distribution with Chi-square test
Fig. 3
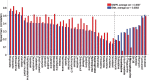
Drug-specific comparison between the baseline GM4 and best GMP6 predictions, based on average Spearman correlation, for 47 cytotoxic drugs. The dotted vertical line distinguishes drugs with improved prediction using proteomics data (left-hand side). The dotted horizontal line indicates well-predicted drugs (correlation ≥ 0.5). Boldfacing marks the example drugs selected for further study (Supplementary Table S2). MoA details are shown in Supplementary Figure S6B
Fig. 4.
Ranked cell-specific prediction of (A) doxorubicin and (B) midostaurin response using the baseline GM4 and best GMP6 model. Value x = 0 is set to equal rank for both the measured and predicted drug responses. Asterisk indicates the case closer to the measured rank for a particular cell line. Color bar quantifies the measured cell line drug sensitivity responses (_GI_50). ROC-AUC calculation was based on quantile value of 0.55
Fig. 5.
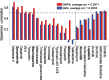
Drug-specific comparison between baseline GM4 and best GMP6 predictions, based on average Spearman correlation, for 24 targeted compounds. The dotted vertical line distinguishes compounds with improved prediction using proteomics data (left-hand side). The dotted horizontal line indicates well-predicted drugs (correlation ≥ 0.5). Boldfacing marks the example compounds selected for further study (Supplementary Table S3). Primary drug target details are available in Supplementary Figure S9B
Fig. 6.
Correlation between measured lapatinib response and ERBB2 protein abundance (available in RPPA only) across all 58 cell lines (solid black line), and in breast and ovarian cancer cells (dotted line). ERBB2 is a known molecular predictor of lapatinib response in breast cancer patients. Statistical significance was assessed with two-sided _t_-test
Similar articles
- Proteomic Approaches for Biomarker Panels in Cancer.
Tanase C, Albulescu R, Neagu M. Tanase C, et al. J Immunoassay Immunochem. 2016;37(1):1-15. doi: 10.1080/15321819.2015.1116009. J Immunoassay Immunochem. 2016. PMID: 26565430 Review. - Topological integration of RPPA proteomic data with multi-omics data for survival prediction in breast cancer via pathway activity inference.
Kim TR, Jeong HH, Sohn KA. Kim TR, et al. BMC Med Genomics. 2019 Jul 11;12(Suppl 5):94. doi: 10.1186/s12920-019-0511-x. BMC Med Genomics. 2019. PMID: 31296204 Free PMC article. - Modeling gene-wise dependencies improves the identification of drug response biomarkers in cancer studies.
Nikolova O, Moser R, Kemp C, Gönen M, Margolin AA. Nikolova O, et al. Bioinformatics. 2017 May 1;33(9):1362-1369. doi: 10.1093/bioinformatics/btw836. Bioinformatics. 2017. PMID: 28082455 Free PMC article. - Evaluation of reverse phase protein array (RPPA)-based pathway-activation profiling in 84 non-small cell lung cancer (NSCLC) cell lines as platform for cancer proteomics and biomarker discovery.
Ummanni R, Mannsperger HA, Sonntag J, Oswald M, Sharma AK, König R, Korf U. Ummanni R, et al. Biochim Biophys Acta. 2014 May;1844(5):950-9. doi: 10.1016/j.bbapap.2013.11.017. Epub 2013 Dec 19. Biochim Biophys Acta. 2014. PMID: 24361481 - The molecular make-up of a tumour: proteomics in cancer research.
Kolch W, Mischak H, Pitt AR. Kolch W, et al. Clin Sci (Lond). 2005 May;108(5):369-83. doi: 10.1042/CS20050006. Clin Sci (Lond). 2005. PMID: 15831087 Review.
Cited by
- Using proteomics to advance the search for potential biomarkers for preeclampsia: A systematic review and meta-analysis.
Nguyen TPH, Patrick CJ, Parry LJ, Familari M. Nguyen TPH, et al. PLoS One. 2019 Apr 5;14(4):e0214671. doi: 10.1371/journal.pone.0214671. eCollection 2019. PLoS One. 2019. PMID: 30951540 Free PMC article. - Predicting drug sensitivity of cancer cells based on DNA methylation levels.
Miranda SP, Baião FA, Fleck JL, Piccolo SR. Miranda SP, et al. PLoS One. 2021 Sep 10;16(9):e0238757. doi: 10.1371/journal.pone.0238757. eCollection 2021. PLoS One. 2021. PMID: 34506489 Free PMC article. - Learning with multiple pairwise kernels for drug bioactivity prediction.
Cichonska A, Pahikkala T, Szedmak S, Julkunen H, Airola A, Heinonen M, Aittokallio T, Rousu J. Cichonska A, et al. Bioinformatics. 2018 Jul 1;34(13):i509-i518. doi: 10.1093/bioinformatics/bty277. Bioinformatics. 2018. PMID: 29949975 Free PMC article. - Systems Biology Approaches Toward Understanding Primary Mitochondrial Diseases.
Maldonado EM, Taha F, Rahman J, Rahman S. Maldonado EM, et al. Front Genet. 2019 Feb 1;10:19. doi: 10.3389/fgene.2019.00019. eCollection 2019. Front Genet. 2019. PMID: 30774647 Free PMC article. Review. - Transforming Diagnosis and Therapeutics Using Cancer Genomics.
Mehmood S, Aslam S, Dilshad E, Ismail H, Khan AN. Mehmood S, et al. Cancer Treat Res. 2023;185:15-47. doi: 10.1007/978-3-031-27156-4_2. Cancer Treat Res. 2023. PMID: 37306902
References
- Aittokallio T. (2010) Dealing with missing values in large-scale studies: microarray data imputation and beyond. Brief. Bioinformatics, 11, 253–264. - PubMed
- Ammad-Ud-Din M. et al. (2016) Drug response prediction by inferring pathway-response associations with kernelized Bayesian matrix factorization. Bioinformatics, 32, i455–i463. - PubMed
- Bhadra S. et al. (2017) Multi-view kernel completion. Mach. Learn., 106, 713–739.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources