Transient receptor potential channel 6 regulates abnormal cardiac S-nitrosylation in Duchenne muscular dystrophy - PubMed (original) (raw)
Transient receptor potential channel 6 regulates abnormal cardiac S-nitrosylation in Duchenne muscular dystrophy
Heaseung Sophia Chung et al. Proc Natl Acad Sci U S A. 2017.
Abstract
Duchenne muscular dystrophy (DMD) is an X-linked disorder with dystrophin loss that results in skeletal and cardiac muscle weakening and early death. Loss of the dystrophin-sarcoglycan complex delocalizes nitric oxide synthase (NOS) to alter its signaling, and augments mechanosensitive intracellular Ca2+ influx. The latter has been coupled to hyperactivation of the nonselective cation channel, transient receptor potential canonical channel 6 (Trpc6), in isolated myocytes. As Ca2+ also activates NOS, we hypothesized that Trpc6 would help to mediate nitric oxide (NO) dysregulation and that this would be manifest in increased myocardial S-nitrosylation, a posttranslational modification increasingly implicated in neurodegenerative, inflammatory, and muscle disease. Using a recently developed dual-labeling proteomic strategy, we identified 1,276 S-nitrosylated cysteine residues [S-nitrosothiol (SNO)] on 491 proteins in resting hearts from a mouse model of DMD (dmdmdx:utrn+/-). These largely consisted of mitochondrial proteins, metabolic regulators, and sarcomeric proteins, with 80% of them also modified in wild type (WT). S-nitrosylation levels, however, were increased in DMD. Genetic deletion of Trpc6 in this model (dmdmdx:utrn+/-:trpc6-/-) reversed ∼70% of these changes. Trpc6 deletion also ameliorated left ventricular dilation, improved cardiac function, and tended to reduce fibrosis. Furthermore, under catecholamine stimulation, which also increases NO synthesis and intracellular Ca2+ along with cardiac workload, the hypernitrosylated state remained as it did at baseline. However, the impact of Trpc6 deletion on the SNO proteome became less marked. These findings reveal a role for Trpc6-mediated hypernitrosylation in dmdmdx:utrn+/- mice and support accumulating evidence that implicates nitrosative stress in cardiac and muscle disease.
Keywords: Duchenne muscular dystrophy; Trpc6; mass spectrometry; nitric oxide synthase signaling; protein S-nitrosylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
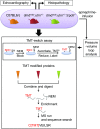
Fig. 1.
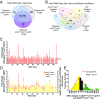
Multiple experimental approaches were employed to search for a linkage between Trpc6, nitrosative stress, and the pathobiology of DMD in the dmdmdx:utrn+/− mice. To capture protein S-nitrosylation, by an MS-coupled dual-labeling strategy, protein mouse heart lysates were labeled with multiplex cys- or iodoTMT6. Samples processed in the absence of ascorbate served as negative controls. Modified proteins were digested, enriched, desalted, and analyzed using MS. C[TMT]VEILSR, an example of TMT-labeled peptides; SH, free cysteine; S-NEM, cysteine blocked with N-ethylmaleimide (NEM).
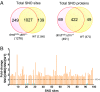
Fig. 2.
Hypernitrosylation of sites found in dmdmdx:utrn+/− cardiac proteins. (A) Bioinformatics identified 1,276 S-nitrosylated residues in dmdmdx:utrn+/− cardiac proteins, and these individual sites (Left) and related proteins (Right) displayed marked commonality between the WT and dmdmdx:utrn+/−. (B) Fold change (FC) in S-nitrosylation level at shared sites between WT and dmdmdx:utrn+/− that exceeded the coefficient of variation (either higher than 1.64 or lower than 0.36) and was detected in at least two biological replicates. Of these 98 sites, the clear majority were more frequently detected in dmdmdx:utrn+/− than in WT [hypernitrosylated (orange), hyponitrosylated (blue)].
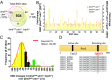
Fig. 3.
Trpc6 deletion reverses many SNO changes in dmdmdx:utrn+/−. (A) Three-way comparison of S-nitrosylation between WT, dmdmdx:utrn+/−, and dmdmdx:utrn+/−:trpc6−/−. More than 78% of sites were detected in all three models. (B) Fold change in S-nitrosylation level detected at a given site comparing dmdmdx:utrn+/− with dmdmdx:utrn+/−:trpc6−/−. Most the sites had less S-nitrosylation detected if Trpc6 was genetically deleted. (C) Histogram of the frequency of percent change in S-nitrosylation between dmdmdx:utrn+/− and dmdmdx:utrn+/−:trpc6−/−. The average of SNO changes in dmdmdx:utrn+/−:trpc6−/− over dmdmdx:utrn+/− was −46.95%; 70% were reversed by 30% or more with Trpc6 deletion (yellow), and 10 sites were increased further by 30% or more (green). The mean of best-fit Gaussian distribution was −60.49%. (D) peroxiredoxin-1 was selected as an example of a potential therapeutic target of Trpc6-dependent SNO proteins. The Cys173 of peroxiredoxin-1 is highly conserved across species, and this residue reacts with Cys52 to form a dimer, which affects the enzymatic activity of peroxiredoxin-1. The Cys173 residue was S-nitrosylated 1.9-fold more in dmdmdx:utrn+/− than control and was restored to normal by >43% in dmdmdx:utrn+/−:trpc6−/−.
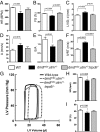
Fig. 4.
LV dilatation increases preload to depress dmdmdx:utrn+/− cardiac function. (A–F) In vivo cardiac function assessed by echocardiography. A, atrial contraction tissue velocity; BPM, beats per minute; E/A, early-to-atrial filling ratio; FS, fractional shortening; HR, heart rate; IVRT, isovolumic ventricular relaxation time; LVID;d, LV diastolic dimension. Pathological changes in all parameters were restored with gene deletion of Trpc6. (WT, n = 8; dmdmdx:utrn+/− dystrophy model, n = 6; and dmdmdx:utrn+/−:trpc6−/− dystrophy model lacking Trpc6, n = 4.) (G–I) Representative pressure-volume loop traces and summary data at rest. The HR and ejection fraction (EF) were reduced in dmdmdx:utrn+/− and restored by Trpc6 deletion. (n = 13, n = 19, and n = 17 for the three groups, respectively.)
Fig. 5.
Trpc6 deletion reverses fibrosis in dmdmdx:utrn+/− hearts. (A) LV myocardium stained with Masson’s trichrome shows increased fibrosis in dmdmdx:utrn+/− mice. This tended to decline with Trpc6 deletion. (B) Summary data (one-way ANOVA, with Tukey’s post hoc test; n = 5–7 per group.) (C) Gene expression for collagen types 1A and 3A (col1a2 and col3a1), ctgf, fn1, and spp1. Most were elevated in dmdmdx:utrn+/−, with Ctgf declining in dmdmdx:utrn+/−:trpc6−/− (one-way ANOVA, with Tukey’s post hoc test, for col1a2 and fn1; Kruskal–Wallis test for col3a1, ctgf, and spp1; n = 5–7 per group.) A.U., arbitrary unit.
Fig. 6.
Influence of epinephrine (Epi) stimulation on SNO proteome in three experimental groups. (A) Venn diagram shows total number of SNO sites identified in Epi-stressed control, dmdmdx:utrn+/− and dmdmdx:utrn+/−:trpc6−/−. (B) Four-way comparison of SNO sites observed in WT and dmdmdx:utrn+/− myocardium ± Epi. (C) Fold change in S-nitrosylation level comparing dmdmdx:utrn+/− with WT. Overall, SNO changes were increased at similar sites in dmdmdx:utrn+/− in the presence of Epi. (D) Fold change in S-nitrosylation level comparing dmdmdx:utrn+/− (red) with dmdmdx:utrn+/−:trpc6−/− (yellow). While some S-nitrosylation levels declined in individual sites, the overall effect was more modest than observed without Epi. (E) Histogram of the relative percent change in S-nitrosylation between dmdmdx:utrn+/− and dmdmdx:utrn+/−:trpc6−/−. Among 90 hyper-SNO sites, 44% were reversed by 30% or more with Trpc6 deletion (yellow), and 22 sites were increased further by 30% or more (green).
Fig. 7.
Bioinformatics analysis provided plausible associations between selected SNO proteins and fibrosis pathways. Proteins that showed SNO change in dmdmdx:utrn+/− were analyzed. Filled shapes indicated the following: proteins where a (or most of the) site(s) was/were hyper-SNO in dmdmdx:utrn+/− (dark orange); proteins where a (or most of the) site(s) was/were hypo-SNO in dmdmdx:utrn+/− (dark green); hyper-SNO or hypo-SNO, but the change did not exceed the average coefficient of variation (light orange or light green); some sites were hyper-SNO and others were hypo-SNO on that protein (mixed orange/green); fibrosis markers confirmed in the level of transcriptome (no filling). Letters in blue indicate SNO on (a) site(s) of the protein was/were reversed with Trpc6-deletion. Of the enriched functional categories, networks of myofilament proteins, multiple isoforms of Prdxs and ILK were selected as being of particular interest. In addition, based on our study on fibrosis, fibrotic markers were included for internetwork generation. Proteins where this network analysis provided the associations between the SNO genes that might affect fibrosis pathways directly/indirectly in DMD pathophysiology are shown.
Similar articles
- Hyperactive adverse mechanical stress responses in dystrophic heart are coupled to transient receptor potential canonical 6 and blocked by cGMP-protein kinase G modulation.
Seo K, Rainer PP, Lee DI, Hao S, Bedja D, Birnbaumer L, Cingolani OH, Kass DA. Seo K, et al. Circ Res. 2014 Feb 28;114(5):823-32. doi: 10.1161/CIRCRESAHA.114.302614. Epub 2014 Jan 21. Circ Res. 2014. PMID: 24449818 Free PMC article. - Pharmacological TRPC6 inhibition improves survival and muscle function in mice with Duchenne muscular dystrophy.
Lin BL, Shin JY, Jeffreys WP, Wang N, Lukban CA, Moorer MC, Velarde E, Hanselman OA, Kwon S, Kannan S, Riddle RC, Ward CW, Pullen SS, Filareto A, Kass DA. Lin BL, et al. JCI Insight. 2022 Oct 10;7(19):e158906. doi: 10.1172/jci.insight.158906. JCI Insight. 2022. PMID: 36099033 Free PMC article. - Intracellular calcium handling in ventricular myocytes from mdx mice.
Williams IA, Allen DG. Williams IA, et al. Am J Physiol Heart Circ Physiol. 2007 Feb;292(2):H846-55. doi: 10.1152/ajpheart.00688.2006. Epub 2006 Sep 29. Am J Physiol Heart Circ Physiol. 2007. PMID: 17012353 - Muscle damage in mdx (dystrophic) mice: role of calcium and reactive oxygen species.
Whitehead NP, Yeung EW, Allen DG. Whitehead NP, et al. Clin Exp Pharmacol Physiol. 2006 Jul;33(7):657-62. doi: 10.1111/j.1440-1681.2006.04394.x. Clin Exp Pharmacol Physiol. 2006. PMID: 16789936 Review. - What has the mdx mouse model of Duchenne muscular dystrophy contributed to our understanding of this disease?
Manning J, O'Malley D. Manning J, et al. J Muscle Res Cell Motil. 2015 Apr;36(2):155-67. doi: 10.1007/s10974-015-9406-4. Epub 2015 Feb 11. J Muscle Res Cell Motil. 2015. PMID: 25669899 Review.
Cited by
- Gain-of-function, focal segmental glomerulosclerosis Trpc6 mutation minimally affects susceptibility to renal injury in several mouse models.
Brown BJ, Boekell KL, Stotter BR, Talbot BE, Schlondorff JS. Brown BJ, et al. PLoS One. 2022 Aug 1;17(8):e0272313. doi: 10.1371/journal.pone.0272313. eCollection 2022. PLoS One. 2022. PMID: 35913909 Free PMC article. - Trpc6 knockout protects against renal fibrosis by restraining the CN‑NFAT2 signaling pathway in T2DM mice.
Sun R, Han M, Liu Y, Su Y, Shi Q, Huang L, Kong L, Li W, Li W. Sun R, et al. Mol Med Rep. 2024 Jan;29(1):13. doi: 10.3892/mmr.2023.13136. Epub 2023 Dec 1. Mol Med Rep. 2024. PMID: 38038121 Free PMC article. - Role of TRPC3 in Right Ventricular Dilatation under Chronic Intermittent Hypoxia in 129/SvEv Mice.
Park DY, Heo W, Kang M, Ahn T, Kim D, Choi A, Birnbaumer L, Cho HJ, Kim JY. Park DY, et al. Int J Mol Sci. 2023 Jul 10;24(14):11284. doi: 10.3390/ijms241411284. Int J Mol Sci. 2023. PMID: 37511045 Free PMC article. - In vivo selective inhibition of TRPC6 by antagonist BI 749327 ameliorates fibrosis and dysfunction in cardiac and renal disease.
Lin BL, Matera D, Doerner JF, Zheng N, Del Camino D, Mishra S, Bian H, Zeveleva S, Zhen X, Blair NT, Chong JA, Hessler DP, Bedja D, Zhu G, Muller GK, Ranek MJ, Pantages L, McFarland M, Netherton MR, Berry A, Wong D, Rast G, Qian HS, Weldon SM, Kuo JJ, Sauer A, Sarko C, Moran MM, Kass DA, Pullen SS. Lin BL, et al. Proc Natl Acad Sci U S A. 2019 May 14;116(20):10156-10161. doi: 10.1073/pnas.1815354116. Epub 2019 Apr 26. Proc Natl Acad Sci U S A. 2019. PMID: 31028142 Free PMC article. - Organic mercury solid phase chemoselective capture for proteomic identification of S-nitrosated proteins and peptides.
Doulias PT, Tenopoulou M, Zakopoulos I, Ischiropoulos H. Doulias PT, et al. Nitric Oxide. 2021 Dec 1;117:1-6. doi: 10.1016/j.niox.2021.09.004. Epub 2021 Sep 15. Nitric Oxide. 2021. PMID: 34536587 Free PMC article.
References
- Davies KE, Nowak KJ. Molecular mechanisms of muscular dystrophies: Old and new players. Nat Rev Mol Cell Biol. 2006;7:762–773. - PubMed
- Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. - PubMed
- Hermans MCE, et al. Hereditary muscular dystrophies and the heart. Neuromuscul Disord. 2010;20:479–492. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 HL112730/HL/NHLBI NIH HHS/United States
- R01 HL119012/HL/NHLBI NIH HHS/United States
- T32 HL007227/HL/NHLBI NIH HHS/United States
- R35 HL135827/HL/NHLBI NIH HHS/United States
- F32 HL010026/HL/NHLBI NIH HHS/United States
- R01 HL131358/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous