Mechanism of a Class C Radical S-Adenosyl-l-methionine Thiazole Methyl Transferase - PubMed (original) (raw)
. 2017 Dec 27;139(51):18623-18631.
doi: 10.1021/jacs.7b10203. Epub 2017 Dec 15.
Affiliations
- PMID: 29190095
- PMCID: PMC5748327
- DOI: 10.1021/jacs.7b10203
Mechanism of a Class C Radical S-Adenosyl-l-methionine Thiazole Methyl Transferase
Zhengan Zhang et al. J Am Chem Soc. 2017.
Abstract
The past decade has seen the discovery of four different classes of radical S-adenosylmethionine (rSAM) methyltransferases that methylate unactivated carbon centers. Whereas the mechanism of class A is well understood, the molecular details of methylation by classes B-D are not. In this study, we present detailed mechanistic investigations of the class C rSAM methyltransferase TbtI involved in the biosynthesis of the potent thiopeptide antibiotic thiomuracin. TbtI C-methylates a Cys-derived thiazole during posttranslational maturation. Product analysis demonstrates that two SAM molecules are required for methylation and that one SAM (SAM1) is converted to 5'-deoxyadenosine and the second SAM (SAM2) is converted to S-adenosyl-l-homocysteine (SAH). Isotope labeling studies show that a hydrogen is transferred from the methyl group of SAM2 to the 5'-deoxyadenosine of SAM1 and the other two hydrogens of the methyl group of SAM2 appear in the methylated product. In addition, a hydrogen appears to be transferred from the β-position of the thiazole to the methyl group in the product. We also show that the methyl protons in the product can exchange with solvent. A mechanism consistent with these observations is presented that differs from other characterized radical SAM methyltransferases.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Figure 1
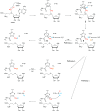
Product analysis for the methyl transfer reaction catalyzed by TbtI. (a) Structure of thiomuracin A1. The methyl group that is introduced by TbtI is shown in red. (b) Regioselective methylation of the thiazole at position 4 of the TbtA core peptide by TbtI. The Val-Gly-Ala sequence at the N-terminus of peptide 1 originates from the leader peptide. (c) Time-dependent formation of the methylated TbtA peptide, 5′-dA, and SAH. Reaction mixtures included the following components: TbtA hexazole (50 μM), [4Fe–4S] cluster reconstituted TbtI (10 μM), SAM (1 mM), flavodoxin (10 μM), flavodoxin reductase (10 μM), NAPDH (2 mM) in reaction buffer (50 mM Tris–HCl, pH 7.5).
Figure 2
Proposed potential reaction mechanisms for TbtI. 5′-dA• is generated from SAM1 in a process mediated by the reduced [4Fe–4S] cluster. The 5′-dA• then abstracts a hydrogen atom from the methyl group of SAM2, and the resulting radical adds to the thiazole. An active site base deprotonates radical 3, leading to the elimination of SAH, yielding radical 4. Three different pathways (a–c) can provide the product and reset the enzyme. In pathway a, the radical is reduced to the anion 5 and protonated. In pathway b, radical 4 abstracts a hydrogen atom from an active site amino acid (X–H). X–H could be the protonated base B–H. In pathway c, radical 4 abstracts a hydrogen atom from the methyl group of 5′-dA. Ade = adenine.
Figure 3
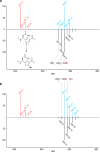
ESI mass spectra to investigate the origin of the methyl protons. 13C- and 15N-depleted substrate peptides were obtained as described (see the Experimental Section). (a) Spectra showing the doubly charged ion for the hexazole-containing core peptide substrate 1 (red) and the methylated product obtained with CD3-SAM in H2O after 1 h (blue) or 16 h (black) reactions. (b) Spectra showing substrate (red) and product obtained with CD3-SAM in D2O (blue) and CD3-5′,5′,4′,3′-D4-SAM in D2O (black) after 1 h. See
Table S2
for calculated and observed masses.
Figure 4
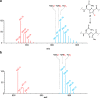
A solvent exchangeable proton migrates from the β-position of thiazole 4 to the methyl group in the product. (a) Spectra showing the doubly charged core peptide of the TbtA hexazole substrate 2 that is deuterated at the β-carbon of each thiazole (red) and the corresponding product obtained with CD3-SAM in D2O (blue) in a single turnover reaction. (b) Spectra showing the doubly charged core peptide of the TbtA hexazole substrate 1 (red) and the corresponding product obtained with CD3-SAM in D2O (blue) in a TbtI-catalyzed single turnover reaction. See
Table S2
for calculated and observed masses.
Similar articles
- Reconstitution and Substrate Specificity of the Radical S-Adenosyl-methionine Thiazole C-Methyltransferase in Thiomuracin Biosynthesis.
Mahanta N, Zhang Z, Hudson GA, van der Donk WA, Mitchell DA. Mahanta N, et al. J Am Chem Soc. 2017 Mar 29;139(12):4310-4313. doi: 10.1021/jacs.7b00693. Epub 2017 Mar 21. J Am Chem Soc. 2017. PMID: 28301141 Free PMC article. - The Catalytic Mechanism of the Class C Radical S-Adenosylmethionine Methyltransferase NosN.
Ding W, Li Y, Zhao J, Ji X, Mo T, Qianzhu H, Tu T, Deng Z, Yu Y, Chen F, Zhang Q. Ding W, et al. Angew Chem Int Ed Engl. 2017 Mar 27;56(14):3857-3861. doi: 10.1002/anie.201609948. Epub 2017 Jan 23. Angew Chem Int Ed Engl. 2017. PMID: 28112859 - GenK-catalyzed C-6' methylation in the biosynthesis of gentamicin: isolation and characterization of a cobalamin-dependent radical SAM enzyme.
Kim HJ, McCarty RM, Ogasawara Y, Liu YN, Mansoorabadi SO, LeVieux J, Liu HW. Kim HJ, et al. J Am Chem Soc. 2013 Jun 5;135(22):8093-6. doi: 10.1021/ja312641f. Epub 2013 May 21. J Am Chem Soc. 2013. PMID: 23679096 Free PMC article. - Radical-mediated enzymatic methylation: a tale of two SAMS.
Zhang Q, van der Donk WA, Liu W. Zhang Q, et al. Acc Chem Res. 2012 Apr 17;45(4):555-64. doi: 10.1021/ar200202c. Epub 2011 Nov 18. Acc Chem Res. 2012. PMID: 22097883 Free PMC article. Review. - Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation.
Bauerle MR, Schwalm EL, Booker SJ. Bauerle MR, et al. J Biol Chem. 2015 Feb 13;290(7):3995-4002. doi: 10.1074/jbc.R114.607044. Epub 2014 Dec 4. J Biol Chem. 2015. PMID: 25477520 Free PMC article. Review.
Cited by
- Docking and MM study of non-structural protein (NS5) of Japanese Encephalitis Virus (JEV) with some derivatives of adenosyl.
Tiwari RK, Pandey V, Srivastava H, Srivastava AK, Pandey V. Tiwari RK, et al. Front Chem. 2023 Nov 27;11:1258764. doi: 10.3389/fchem.2023.1258764. eCollection 2023. Front Chem. 2023. PMID: 38090351 Free PMC article. - Methanogenesis marker protein 10 (Mmp10) from Methanosarcina acetivorans is a radical _S_-adenosylmethionine methylase that unexpectedly requires cobalamin.
Radle MI, Miller DV, Laremore TN, Booker SJ. Radle MI, et al. J Biol Chem. 2019 Aug 2;294(31):11712-11725. doi: 10.1074/jbc.RA119.007609. Epub 2019 May 20. J Biol Chem. 2019. PMID: 31113866 Free PMC article. - Functional Diversity of HemN-like Proteins.
Cheng J, Liu WQ, Zhu X, Zhang Q. Cheng J, et al. ACS Bio Med Chem Au. 2022 Jan 18;2(2):109-119. doi: 10.1021/acsbiomedchemau.1c00058. eCollection 2022 Apr 20. ACS Bio Med Chem Au. 2022. PMID: 37101745 Free PMC article. Review. - Formation and Cleavage of C-C Bonds by Enzymatic Oxidation-Reduction Reactions.
Guengerich FP, Yoshimoto FK. Guengerich FP, et al. Chem Rev. 2018 Jul 25;118(14):6573-6655. doi: 10.1021/acs.chemrev.8b00031. Epub 2018 Jun 22. Chem Rev. 2018. PMID: 29932643 Free PMC article. Review. - A radical S-adenosyl-L-methionine enzyme and a methyltransferase catalyze cyclopropane formation in natural product biosynthesis.
Jin WB, Wu S, Jian XH, Yuan H, Tang GL. Jin WB, et al. Nat Commun. 2018 Jul 17;9(1):2771. doi: 10.1038/s41467-018-05217-1. Nat Commun. 2018. PMID: 30018376 Free PMC article.
References
- Takusagawa F.; Fujioka M.; Spies A.; Schowen R. L. In Comprehensive Biological Catalysis; Sinnott M., Ed.; Academic Press: New York, 1998; p 1.
- Williamson J. M.; Inamine E.; Wilson K. E.; Douglas A. W.; Liesch J. M.; Albers-Schönberg G. J. Biol. Chem. 1985, 260, 4637. - PubMed
- Houck D. R.; Kobayashi K.; Williamson J. M.; Floss H. G. J. Am. Chem. Soc. 1986, 108, 5365.10.1021/ja00277a063. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous