Transcriptome profiling analysis of senescent gingival fibroblasts in response to Fusobacterium nucleatum infection - PubMed (original) (raw)
Transcriptome profiling analysis of senescent gingival fibroblasts in response to Fusobacterium nucleatum infection
Sun-Hee Ahn et al. PLoS One. 2017.
Erratum in
- Correction: Transcriptome profiling analysis of senescent gingival fibroblasts in response to Fusobacterium nucleatum infection.
Ahn SH, Cheon S, Park C, Lee JH, Lee SW, Lee TH. Ahn SH, et al. PLoS One. 2018 Jan 9;13(1):e0191209. doi: 10.1371/journal.pone.0191209. eCollection 2018. PLoS One. 2018. PMID: 29315353 Free PMC article.
Abstract
Periodontal disease is caused by dental plaque biofilms. Fusobacterium nucleatum is an important periodontal pathogen involved in the development of bacterial complexity in dental plaque biofilms. Human gingival fibroblasts (GFs) act as the first line of defense against oral microorganisms and locally orchestrate immune responses by triggering the production of reactive oxygen species and pro-inflammatory cytokines (IL-6 and IL-8). The frequency and severity of periodontal diseases is known to increase in elderly subjects. However, despite several studies exploring the effects of aging in periodontal disease, the underlying mechanisms through which aging affects the interaction between F. nucleatum and human GFs remain unclear. To identify genes affected by infection, aging, or both, we performed an RNA-Seq analysis using GFs isolated from a single healthy donor that were passaged for a short period of time (P4) 'young GFs' or for longer period of time (P22) 'old GFs', and infected or not with F. nucleatum. Comparing F. nucleatum-infected and uninfected GF(P4) cells the differentially expressed genes (DEGs) were involved in host defense mechanisms (i.e., immune responses and defense responses), whereas comparing F. nucleatum-infected and uninfected GF(P22) cells the DEGs were involved in cell maintenance (i.e., TGF-β signaling, skeletal development). Most DEGs in F. nucleatum-infected GF(P22) cells were downregulated (85%) and were significantly associated with host defense responses such as inflammatory responses, when compared to the DEGs in F. nucleatum-infected GF(P4) cells. Five genes (GADD45b, KLF10, CSRNP1, ID1, and TM4SF1) were upregulated in response to F. nucleatum infection; however, this effect was only seen in GF(P22) cells. The genes identified here appear to interact with each other in a network associated with free radical scavenging, cell cycle, and cancer; therefore, they could be potential candidates involved in the aged GF's response to F. nucleatum infection. Further studies are needed to confirm these observations.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Fig 1. RNA-seq analysis pipeline.
Laboratory pipeline for simultaneous depletion of rRNA from prokaryotic and eukaryotic RNA mixtures. The enriched mRNA was used to generate the RNA-Seq libraries.
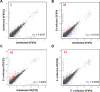
Fig 2. Distribution of RNA-seq data.
All gene expression levels were transformed to base two logarithms. (A) Uninfected GFs (P4) vs. uninfected GFs (P22). (B) Uninfected GFs (P4) vs. _Fusobacterium nucleatum_-infected GFs (P4). (C) Uninfected GFs (P22) vs. F. _nucleatum_-infected GFs (P22). (D) F. _nucleatum_-infected GFs (P4) vs. F. _nucleatum_-infected GFs (P22). The individual Spearman's rank correlation coefficients are shown on each graph. Significant DEGs between the two samples, with an FDR <5% when Benjamini-Hochberg multiple testing adjustment was used, are shown in red dots (upregulated) and blue dots (downregulated).
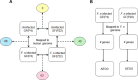
Fig 3. Strategy for the analysis of RNA-seq data.
(A) The RNA-seq data was mapped onto the human genome and the indicated comparisons were conducted. The number of DEGs from each comparison is indicated in the circle. (B) The RNA-seq data were mapped onto the Fusobacterium nucleatum genome.
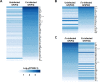
Fig 4. Heat maps for the different gene expression comparisons.
(A) Heat map of the eighty-eight DEGs identified between uninfected GF(P4) and F. _nucleatum_-infected GF(P4) cells. (B) Heat map of the forty DEGs identified between uninfected GF(P22) and _Fusobacterium nucleatum_-infected GF(P22) cells. (C) Heat map of the sixty-two DEGs identified between F. _nucleatum_-infected GF(P4) and F. _nucleatum_-infected GF(P22) cells. Genes were considered to be significantly differentially expressed if they obtained a q < 0.05 using a Benjamini-Hochberg multiple testing adjustment.
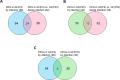
Fig 5. Venn diagram summarizing the overlap analysis of DEGs from the three paired comparisons.
(A) Young GF(P4)-specific response to infection. (B) Aged GF(P22)-specific response to infection. (C) Common genes in young (P4) and aged (P22) GFs in response to infection.
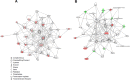
Fig 6. Networks predicted by IPA for the DEGs.
IPA network analysis of the genes from (A) young GF(P4)-specific response to F. nucleatum infection and the genes from (B) aged GF(P22)-specific response to F. nucleatum infection. The network is displayed graphically as nodes (genes). The node color intensity indicates the expression of genes, with red representing upregulation and green representing downregulation. Solid lines and dotted lines indicate direct relationship and indirect relationships, respectively. Red and blue denote upregulated and downregulated DEGs, respectively.
Similar articles
- Caveolin-1 serves as a negative effector in senescent human gingival fibroblasts during Fusobacterium nucleatum infection.
Ahn SH, Cho SH, Song JE, Kim S, Oh SS, Jung S, Cho KA, Lee TH. Ahn SH, et al. Mol Oral Microbiol. 2017 Jun;32(3):236-249. doi: 10.1111/omi.12167. Epub 2016 Jul 31. Mol Oral Microbiol. 2017. PMID: 27315395 - NOX1/2 activation in human gingival fibroblasts by Fusobacterium nucleatum facilitates attachment of Porphyromonas gingivalis.
Ahn SH, Song JE, Kim S, Cho SH, Lim YK, Kook JK, Kook MS, Lee TH. Ahn SH, et al. Arch Microbiol. 2016 Aug;198(6):573-83. doi: 10.1007/s00203-016-1223-7. Epub 2016 Apr 12. Arch Microbiol. 2016. PMID: 27071620 - Fusobacterium nucleatum enters normal human oral fibroblasts in vitro.
Dabija-Wolter G, Cimpan MR, Costea DE, Johannessen AC, Sørnes S, Neppelberg E, Al-Haroni M, Skaug N, Bakken V. Dabija-Wolter G, et al. J Periodontol. 2009 Jul;80(7):1174-83. doi: 10.1902/jop.2009.090051. J Periodontol. 2009. PMID: 19563299 - Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer.
Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H, Maruyama R, Imai K, Yamamoto H, Shinomura Y. Nosho K, et al. World J Gastroenterol. 2016 Jan 14;22(2):557-66. doi: 10.3748/wjg.v22.i2.557. World J Gastroenterol. 2016. PMID: 26811607 Free PMC article. Review. - Fusobacterium nucleatum: an emerging gut pathogen?
Allen-Vercoe E, Strauss J, Chadee K. Allen-Vercoe E, et al. Gut Microbes. 2011 Sep 1;2(5):294-8. doi: 10.4161/gmic.2.5.18603. Epub 2011 Sep 1. Gut Microbes. 2011. PMID: 22067936 Review.
Cited by
- A bibliometric and visualized in oral microbiota and cancer research from 2013 to 2022.
Gu Z, Liu Y. Gu Z, et al. Discov Oncol. 2024 Feb 1;15(1):24. doi: 10.1007/s12672-024-00878-5. Discov Oncol. 2024. PMID: 38302656 Free PMC article. - Creutzfeldt-Jakob Disease: Alterations of Gut Microbiota.
Guo Y, Xu Y, Lin X, Zhen Z, Yi F, Guan H, Shi Q, Sun W, Yang A, Dong X, Wang J. Guo Y, et al. Front Neurol. 2022 Apr 15;13:832599. doi: 10.3389/fneur.2022.832599. eCollection 2022. Front Neurol. 2022. PMID: 35493823 Free PMC article. - Molecular Signatures of Senescence in Periodontitis: Clinical Insights.
Rattanaprukskul K, Xia XJ, Jiang M, Albuquerque-Souza E, Bandyopadhyay D, Sahingur SE. Rattanaprukskul K, et al. J Dent Res. 2024 Jul;103(8):800-808. doi: 10.1177/00220345241255325. Epub 2024 Jun 14. J Dent Res. 2024. PMID: 38877743 Free PMC article. - Oral hygiene might prevent cancer.
Cordero OJ, Varela-Calviño R. Cordero OJ, et al. Heliyon. 2018 Nov 2;4(10):e00879. doi: 10.1016/j.heliyon.2018.e00879. eCollection 2018 Oct. Heliyon. 2018. PMID: 30417145 Free PMC article. Review. - Fusobacterium nucleatum - symbiont, opportunist and oncobacterium.
Brennan CA, Garrett WS. Brennan CA, et al. Nat Rev Microbiol. 2019 Mar;17(3):156-166. doi: 10.1038/s41579-018-0129-6. Nat Rev Microbiol. 2019. PMID: 30546113 Free PMC article. Review.
References
- Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4(1):1–6. doi: 10.1902/annals.1999.4.1.1 . - DOI - PubMed
- Kolenbrander PE. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol. 2000;54:413–37. doi: 10.1146/annurev.micro.54.1.413 . - DOI - PubMed
- van Winkelhoff AJ, Loos BG, van der Reijden WA, van der Velden U. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. Journal of clinical periodontology. 2002;29(11):1023–8. . - PubMed
- Diaz PI, Zilm PS, Rogers AH. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology. 2002;148(Pt 2):467–72. doi: 10.1099/00221287-148-2-467 . - DOI - PubMed
- Signat B, Roques C, Poulet P, Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Current issues in molecular biology. 2011;13(2):25–36. . - PubMed
MeSH terms
Grants and funding
This work was supported by the Bio & Medical Technology Development Program (2016M3A9B6903087) and the Basic Science Research Program (2017R1A2B2005938) of the National Research Foundation (NRF) funded by the Ministry of Science, ICT & Future Planning. Sun Hee Ahn was supported by the Basic Science Research Program (2016R1A6A3A11932468) through the NRF funded by the Ministry of Education and The grant from the World Institute of Kimchi (KE1701-2) to Dr. Jong-Hee Lee. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources