Multilocus DNA barcoding - Species Identification with Multilocus Data - PubMed (original) (raw)
Multilocus DNA barcoding - Species Identification with Multilocus Data
Junning Liu et al. Sci Rep. 2017.
Abstract
Species identification using DNA sequences, known as DNA barcoding has been widely used in many applied fields. Current barcoding methods are usually based on a single mitochondrial locus, such as cytochrome c oxidase subunit I (COI). This type of barcoding method does not always work when applied to species separated by short divergence times or that contain introgressed genes from closely related species. Herein we introduce a more effective multi-locus barcoding framework that is based on gene capture and "next-generation" sequencing. We selected 500 independent nuclear markers for ray-finned fishes and designed a three-step pipeline for multilocus DNA barcoding. We applied our method on two exemplar datasets each containing a pair of sister fish species: Siniperca chuatsi vs. Sini. kneri and Sicydium altum vs. Sicy. adelum, where the COI barcoding approach failed. Both of our empirical and simulated results demonstrated that under limited gene flow and enough separation time, we could correctly identify species using multilocus barcoding method. We anticipate that, as the cost of DNA sequencing continues to fall that our multilocus barcoding approach will eclipse existing single-locus DNA barcoding methods as a means to better understand the diversity of the living world.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
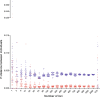
Figure 1
Intra- (red) and interspecific (blue) p-distance of Siniperca chuatsi and Sini. kneri calculated using different number of nuclear loci or COI gene. Scale of distances larger than 0.020 was reduced to fit all data points in the art board.
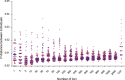
Figure 2
Intra- (red) and interspecific (blue) p-distance of Sicydium altum and Sicy. adelum calculated using different numbers of nuclear loci or COI gene.
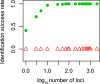
Figure 3
The relationship between number of loci used and success rate of identification between Siniperca chuatsi and Sini. kneri (green dots), and between Sicydium altum and Sicy. adelum (red triangles).
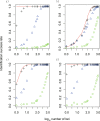
Figure 4
Identification success rate using simulated sequences under different scenarios. (a) migration rate equals zero and divergence time equals 700,000 generations (red line), 100,000 generations (black crosses), 10,000 generations (blue triangles), and 1000 generations (green circles); (b) divergence time equals 10,000 and migration rate equals 0 (red line), 0.000001 (black crosses), 0.00001 (blue triangles) and 0.0001 (green circles); (c) divergence time equals 100,000 and migration rate equals 0 (red line), 0.000001 (black crosses), 0.00001 (blue triangles) and 0.0001 (green circles); (d) divergence time equals 700,000 and migration rate equals 0 (red line), 0.000001 (black crosses), 0.00001 (blue triangles) and 0.0001 (green circles).
Figure 5
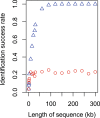
Comparison between success rates of species identification based on a single locus (red circles) and multiple loci (blue triangles). The length of the single locus equals the total length of multiple loci (300 bp each).
Figure 6
A three-step multilocus DNA barcoding pipeline.
Similar articles
- DNA barcoding to fishes: current status and future directions.
Bhattacharya M, Sharma AR, Patra BC, Sharma G, Seo EM, Nam JS, Chakraborty C, Lee SS. Bhattacharya M, et al. Mitochondrial DNA A DNA Mapp Seq Anal. 2016 Jul;27(4):2744-52. doi: 10.3109/19401736.2015.1046175. Epub 2015 Jun 9. Mitochondrial DNA A DNA Mapp Seq Anal. 2016. PMID: 26057011 - DNA barcoding for identification of fish species in the Taiwan Strait.
Bingpeng X, Heshan L, Zhilan Z, Chunguang W, Yanguo W, Jianjun W. Bingpeng X, et al. PLoS One. 2018 Jun 1;13(6):e0198109. doi: 10.1371/journal.pone.0198109. eCollection 2018. PLoS One. 2018. PMID: 29856794 Free PMC article. - DNA barcoding Indian marine fishes.
Lakra WS, Verma MS, Goswami M, Lal KK, Mohindra V, Punia P, Gopalakrishnan A, Singh KV, Ward RD, Hebert P. Lakra WS, et al. Mol Ecol Resour. 2011 Jan;11(1):60-71. doi: 10.1111/j.1755-0998.2010.02894.x. Mol Ecol Resour. 2011. PMID: 21429101 - Multilocus inference of species trees and DNA barcoding.
Mallo D, Posada D. Mallo D, et al. Philos Trans R Soc Lond B Biol Sci. 2016 Sep 5;371(1702):20150335. doi: 10.1098/rstb.2015.0335. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27481787 Free PMC article. Review. - Aspergillus and Penicillium identification using DNA sequences: barcode or MLST?
Peterson SW. Peterson SW. Appl Microbiol Biotechnol. 2012 Jul;95(2):339-44. doi: 10.1007/s00253-012-4165-2. Epub 2012 May 27. Appl Microbiol Biotechnol. 2012. PMID: 22639145 Review.
Cited by
- A high-resolution genome of an euryhaline and eurythermal rhinogoby (Rhinogobius similis Gill 1895).
Hu Y, Lu L, Zhou T, Sarker KK, Huang J, Xia J, Li C. Hu Y, et al. G3 (Bethesda). 2022 Feb 4;12(2):jkab395. doi: 10.1093/g3journal/jkab395. G3 (Bethesda). 2022. PMID: 34792546 Free PMC article. - A comprehensive DNA barcoding reference database for Plecoptera of Switzerland.
Vuataz L, Reding JP, Reding A, Roesti C, Stoffel C, Vinçon G, Gattolliat JL. Vuataz L, et al. Sci Rep. 2024 Mar 15;14(1):6322. doi: 10.1038/s41598-024-56930-5. Sci Rep. 2024. PMID: 38491157 Free PMC article. - Molecular phylogenetic analysis and morphological reassessments of thief ants identify a new potential case of biological invasions.
Sharaf MR, Gotzek D, Guénard B, Fisher BL, Aldawood AS, Al Dhafer HM, Mohamed AA. Sharaf MR, et al. Sci Rep. 2020 Jul 21;10(1):12040. doi: 10.1038/s41598-020-69029-4. Sci Rep. 2020. PMID: 32694527 Free PMC article. - ACDC, a global database of amphibian cytochrome-b sequences using reproducible curation for GenBank records.
van den Burg MP, Herrando-Pérez S, Vieites DR. van den Burg MP, et al. Sci Data. 2020 Aug 13;7(1):268. doi: 10.1038/s41597-020-00598-9. Sci Data. 2020. PMID: 32792559 Free PMC article. - Cox1 barcoding versus multilocus species delimitation: validation of two mite species with contrasting effective population sizes.
Klimov PB, Skoracki M, Bochkov AV. Klimov PB, et al. Parasit Vectors. 2019 Jan 5;12(1):8. doi: 10.1186/s13071-018-3242-5. Parasit Vectors. 2019. PMID: 30611284 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources