Massive Iatrogenic Outbreak of Human Immunodeficiency Virus Type 1 in Rural Cambodia, 2014-2015 - PubMed (original) (raw)
. 2018 May 17;66(11):1733-1741.
doi: 10.1093/cid/cix1071.
Janin Nouhin 1, Du-Ping Zheng 2, Benjamin Roche 3, Allison Black 4, Sophearot Prak 1, Marie Leoz 5, Catherine Gaudy-Graffin 6, Laurent Ferradini 7, Chandara Mom 8, Sovatha Mam 8, Charlotte Gautier 1, Gérard Lesage 6, Sreymom Ken 1, Kerya Phon 1, Alexandra Kerleguer 1, Chunfu Yang 2, William Killam 9, Masami Fujita 7, Chhivun Mean 8, Didier Fontenille 1, Francis Barin 6, Jean-Christophe Plantier 5, Trevor Bedford 4, Artur Ramos 9, Vonthanak Saphonn 10
Affiliations
- PMID: 29211835
- PMCID: PMC5963970
- DOI: 10.1093/cid/cix1071
Massive Iatrogenic Outbreak of Human Immunodeficiency Virus Type 1 in Rural Cambodia, 2014-2015
François Rouet et al. Clin Infect Dis. 2018.
Abstract
Background: In 2014-2015, 242 individuals aged 2-89 years were newly diagnosed with human immunodeficiency virus type 1 (HIV-1) in Roka, a rural commune in Cambodia. A case-control study attributed the outbreak to unsafe injections. We aimed to reconstruct the likely transmission history of the outbreak.
Methods: We assessed in 209 (86.4%) HIV-infected cases the presence of hepatitis C virus (HCV) and hepatitis B virus (HBV). We identified recent infections using antibody (Ab) avidity testing for HIV and HCV. We performed amplification, sequencing, and evolutionary phylogenetic analyses of viral strains. Geographical coordinates and parenteral exposure through medical services provided by an unlicensed healthcare practitioner were obtained from 193 cases and 1499 controls during interviews.
Results: Cases were coinfected with HCV (78.5%) and HBV (12.9%). We identified 79 (37.8%) recent (<130 days) HIV infections. Phylogeny of 202 HIV env C2V3 sequences showed a 198-sample CRF01_AE strains cluster, with time to most recent common ancestor (tMRCA) in September 2013 (95% highest posterior density, August 2012-July 2014), and a peak of 15 infections/day in September 2014. Three geospatial HIV hotspots were discernible in Roka and correlated with high exposure to the practitioner (P = .04). Fifty-nine of 153 (38.6%) tested cases showed recent (<180 days) HCV infections. Ninety HCV NS5B sequences formed 3 main clades, 1 containing 34 subtypes 1b with tMRCA in 2012, and 2 with 51 subtypes 6e and tMRCAs in 2002-2003.
Conclusions: Unsafe injections in Cambodia most likely led to an explosive iatrogenic spreading of HIV, associated with a long-standing and more genetically diverse HCV propagation.
Figures
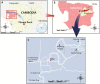
Figure 1.
Geographical location of the iatrogenic human immunodeficiency virus outbreak in the Roka commune, Cambodia, 2014–2015. A, Map showing Cambodia (in yellow) and the Battambang province (in pink). B, Map showing the Roka commune (in red) which is located in the Sangkae district (in yellow). C, Map showing the location of the 6 villages in the Roka commune (7985 inhabitants). Roka village (2338 inhabitants) is indicated by a red circle while the other 5 villages (Ambaeng Thngae [1050 inhabitants], Ta Haen I [1468 inhabitants], Ta Haen II [1050 inhabitants], Pou Batdambang [731 inhabitants], and Chhung Tradak [1348 inhabitants]) are indicated by black circles. Roads are indicated by white lines.
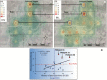
Figure 2.
Human immunodeficiency virus type 1 (HIV-1) env C2V3 sequences. A, Maximum-likelihood phylogenetic tree for HIV-1 env C2V3 sequences from 202 case sequences (indicated in red), 45 Cambodia sequences (blue), and 24 GenBank reference sequences (black). The Roka outbreak–associated CRF01_AE HIV cluster is represented by a red triangle. Four sequences (amplified from National Center for HIV/AIDS, Dermatology and Sexually Transmitted Diseases [NCHADS] patients 171, 184, 185, and 116) from Roka did not group within the outbreak-associated cluster, and were isolated from individuals presenting nonrecent HIV-1 infections and who were negative for HCV and HBV. The unit for the scale bar of 0.05 is the number of nucleotide substitutions per site. GenBank accession numbers for the Roka HIV env C2V3 sequences were KY570019–KY570220. Accession numbers for other Cambodia sequences were KY570221–KY570265. B, Bayesian time-scaled phylogenetic tree of 198 HIV-1 env C2V3 sequences from the Roka HIV cluster. The branch lengths represent the number of substitutions per site per year. The cross marks the mean estimate of the time to the most recent common ancestor with 95% highest posterior densities (HPDs). C, Effective viral population size over time. The Bayesian skyline plot shows the median viral population size over time (black solid line) and 95% HPDs around the estimate (black dashed lines). Expansion model is indicated by a red line. D, Lineage-through-time plot. CLTT indicates the cumulative lineage through time (black solid line); NLTT indicates the noncumulative lineage through time (red solid line). Abbreviations: HBV, hepatitis B virus; HCV, hepatitis C virus; Ne, effective population size.
Figure 3.
Geographic variations in interpolated human immunodeficiency virus (HIV) prevalence (A), frequency of history of injections (INJ.) with the unlicensed practitioner (B) across the surveillance area, and correlation (C). Roka, Ambang Thngae, and Ta Haen I villages are delineated by a dotted rectangle. The practitioner’s house is depicted by a white cross. Each hotspot is indicated by a circle, associated with a number (1, 2, or 3 for HIV) or a letter (a, b, or c regarding history of past injections).
Figure 4.
Hepatitis C virus (HCV) NS5B sequences. A, Maximum-likelihood phylogenetic tree for HCV NS5B sequences from 39 patients with HCV subtype 1b sequences (red), 37 Cambodia sequences (blue), and 29 GenBank reference sequences (black). GenBank accession numbers for the Roka HCV 1b NS5B sequences were KY569650–KY569688. Accession numbers for other Cambodia sequences were KY569689–KY569725. B, Bayesian time-scaled phylogenetic tree of 34 HCV NS5B sequences from the Roka subtype 1b HCV cluster. The branch lengths represent the number of substitutions per site per year. The cross marks the mean estimate of the time to the most recent common ancestor (tMRCA) with 95% highest posterior densities (HPDs). C, Maximum-likelihood phylogenetic tree for HCV NS5B sequences from 51 patients with HCV subtype 6e sequences (red), 18 Cambodia sequences (blue), and 29 GenBank reference sequences (black). The 51 HCV subtype 6e strains were distributed in 2 distinct monophyletic clusters. GenBank accession numbers for the Roka HCV 6e sequences were KY569734–KY569784. Accession numbers for other Cambodia sequences were KY569785–KY569802. D, Bayesian time-scaled phylogenetic tree of 51 HCV NS5B sequences from the Roka genotype 6e clusters (n = 51). The branch lengths represent the number of substitutions per site per year. The mean estimate of the tMRCAs as well as 95% HPDs are indicated with at key internal nodes marked with “X.”
Similar articles
- Phylogenetic Analysis of Spread of Hepatitis C Virus Identified during HIV Outbreak Investigation, Unnao, India.
Mane A, Kasibhatla SM, Vidhate P, Saxena V, Patil S, Rao A, Nirmalkar A, Kulkarni-Kale U, Panda S. Mane A, et al. Emerg Infect Dis. 2022 Apr;28(4):725-733. doi: 10.3201/eid2804.211845. Emerg Infect Dis. 2022. PMID: 35318918 Free PMC article. Review. - Cluster of HIV Infections Attributed to Unsafe Injection Practices--Cambodia, December 1, 2014-February 28, 2015.
Vun MC, Galang RR, Fujita M, Killam W, Gokhale R, Pitman J, Selenic D, Mam S, Mom C, Fontenille D, Rouet F, Vonthanak S; Roka Cluster Investigation Team. Vun MC, et al. MMWR Morb Mortal Wkly Rep. 2016 Feb 19;65(6):142-5. doi: 10.15585/mmwr.mm6506a2. MMWR Morb Mortal Wkly Rep. 2016. PMID: 26890340 - Cluster of HIV Infections Associated With Unsafe Injection Practices in a Rural Village in Cambodia.
Saphonn V, Fujita M, Samreth S, Chan S, Rouet F, Khol V, Mam S, Mom C, Tuot S, Le LV, Ly PS, Ferradini L, Mean CV. Saphonn V, et al. J Acquir Immune Defic Syndr. 2017 Jul 1;75(3):e82-e86. doi: 10.1097/QAI.0000000000001295. J Acquir Immune Defic Syndr. 2017. PMID: 28129255 Free PMC article. No abstract available. - Notes from the Field: Public Health Response to a Human Immunodeficiency Virus Outbreak Associated with Unsafe Injection Practices - Roka Commune, Cambodia, 2016.
Ijeoma UC, Sansam S, Srun S, Vannara H, Sanith S, Sopheap T, Newman RD, Gadde R, Dejana S, Hassani AS, Ly V, Drammeh B, De A, Byrd J, Bock N. Ijeoma UC, et al. MMWR Morb Mortal Wkly Rep. 2018 Feb 2;67(4):135-136. doi: 10.15585/mmwr.mm6704a6. MMWR Morb Mortal Wkly Rep. 2018. PMID: 29389915 Free PMC article. No abstract available. - Rapid assessment of injection practices in Cambodia, 2002.
Vong S, Perz JF, Sok S, Som S, Goldstein S, Hutin Y, Tulloch J. Vong S, et al. BMC Public Health. 2005 Jun 2;5:56. doi: 10.1186/1471-2458-5-56. BMC Public Health. 2005. PMID: 15929800 Free PMC article.
Cited by
- Regulation of nursing professionals in Cambodia and Vietnam: a review of the evolution and key influences.
Fujita N, Matsuoka S, Koto-Shimada K, Ikarashi M, Hazarika I, Zwi AB. Fujita N, et al. Hum Resour Health. 2019 Jul 3;17(1):48. doi: 10.1186/s12960-019-0388-y. Hum Resour Health. 2019. PMID: 31269960 Free PMC article. Review. - Molecular epidemiology of hepatitis C virus in Cambodia during 2016-2017.
Nouhin J, Iwamoto M, Prak S, Dousset JP, Phon K, Heng S, Kerleguer A, Le Paih M, Dussart P, Maman D, Rouet F. Nouhin J, et al. Sci Rep. 2019 May 13;9(1):7314. doi: 10.1038/s41598-019-43785-4. Sci Rep. 2019. PMID: 31086236 Free PMC article. - Phylogenetic Analysis of Spread of Hepatitis C Virus Identified during HIV Outbreak Investigation, Unnao, India.
Mane A, Kasibhatla SM, Vidhate P, Saxena V, Patil S, Rao A, Nirmalkar A, Kulkarni-Kale U, Panda S. Mane A, et al. Emerg Infect Dis. 2022 Apr;28(4):725-733. doi: 10.3201/eid2804.211845. Emerg Infect Dis. 2022. PMID: 35318918 Free PMC article. Review. - Phylogenetics applied to the human immunodeficiency virus type 1 (HIV-1): from the cross-species transmissions to the contact network inferences.
Gräf T, Delatorre E, Bello G. Gräf T, et al. Mem Inst Oswaldo Cruz. 2020 Mar 16;115:e190461. doi: 10.1590/0074-02760190461. eCollection 2020. Mem Inst Oswaldo Cruz. 2020. PMID: 32187328 Free PMC article.
References
- World Health Organization. Global hepatitis report, 2017. Available at: http://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/. Accessed 10 May 2017.
- Vun MC, Galang RR, Fujita M, et al. . Cluster of HIV infections attributed to unsafe injection practices—Cambodia, December 1, 2014–February 28, 2015. MMWR Morb Mortal Wkly Rep 2016; 65:142–5. - PubMed
- Phnom Penh Post. Doctor gets 25 years for HIV outbreak Available at: http://www.phnompenhpost.com/national/doctor-gets-25-years-hiv-outbreak. Accessed 10 May 2017.
Publication types
MeSH terms
Grants and funding
- 001/WHO_/World Health Organization/International
- CC999999/ImCDC/Intramural CDC HHS/United States
- S10 OD020069/OD/NIH HHS/United States
- U2G GH000989/GH/CGH CDC HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases