3D microniches reveal the importance of cell size and shape - PubMed (original) (raw)
3D microniches reveal the importance of cell size and shape
Min Bao et al. Nat Commun. 2017.
Abstract
Geometrical cues have been shown to alter gene expression and differentiation on 2D substrates. However, little is known about how geometrical cues affect cell function in 3D. One major reason for this lack of understanding is rooted in the difficulties of controlling cell geometry in a complex 3D setting and for long periods of culture. Here, we present a robust method to control cell volume and shape of individual human mesenchymal stem cells (hMSCs) inside 3D microniches with a range of different geometries (e.g., cylinder, triangular prism, cubic, and cuboid). We find that the actin filaments, focal adhesions, nuclear shape, YAP/TAZ localization, cell contractility, nuclear accumulation of histone deacetylase 3, and lineage selection are all sensitive to cell volume. Our 3D microniches enable fundamental studies on the impact of biophysical cues on cell fate, and have potential applications in investigating how multicellular architectures organize within geometrically well-defined 3D spaces.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures
Fig. 1
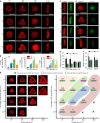
3D microniche preparation and single hMSC encapsulation. a Schematic of the method to encapsulate single cells in a 3D microniche. b Fluorescence image shows nuclear staining of single cells encapsulated in a 3D microniche with cylindrical geometries at different cell densities (2500 and 10,000 cells cm−2), scale bar: 100 µm. c Cell encapsulation efficiency at different cell densities in the 3D microenvironment with cylindrical geometry. d Quantification of cell viability (by live/dead staining) after 1 and 3 days of culture in a 3D microniche with different geometries; n ≥4 regions of interest (ROI) with a total of 80–100 cells analyzed. e Side view and fluorescent heat maps of actin organization in a microenvironment with and without lid, red: F-actin, scale bar: 20 µm. f 3D organization of actin cytoskeleton in a microenvironment with and without lid, red: F-actin, blue: nuclear, scale bar: 20 µm. g Quantification of cell volume after 24-h culture in a microenvironment with and without lid; n = 50–60 cells analyzed for each data point. The microniche volume was controlled by changing the height (from 7 to 30 µm), with a constant value for project area (400 µm2). Data are shown as mean ± s.d. for all panels, and *P < 0.05, **P < 0.01 (ANOVA using a Tukey post-test), compared to theoretical niche volume. Microniches with heights 23, 12, 9, and 7 µm are denoted as V 1, V 2, V 3, and V 4, respectively
Fig. 2
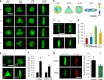
F-actin filaments formation and polymerization in a 3D microniche. a Representative images of F-actin staining for hMSCs with different cell volumes and cell geometries after 24 h. b Quantification of the number of cells forming stress fibers in a 3D microniche with different sizes and geometries; n = 50–60 cells analyzed for each data point. c Immunofluorescence images of F-actin and G-actin for hMSCs with different volumes after 12 h. d Quantification of F- and G-actin levels 12 h after seeding in 3D microniches with different volumes. Total integrated fluorescence of phalloidin (F-actin) and DNaseI (G-actin) was normalized to the fluorescence of V 3 cells; n = 40–45 cells analyzed for each data point. e Immunofluorescence images of F-actin and G-actin for hMSCs with different geometries with V 3 volume after 12 h. f Comparison of normalized mean F- and G-actin intensity in cells with different shapes (cylinder and triangular prism) and aspect ratios (cubic and cuboid); n = 40–45 cells analyzed for each data point. g Left: F-actin staining for single hMSCs cultured in 3D microniches with different volumes. Representative cells were selected for each condition. Right: quantification of the number of cells forming stress fibers in 3D microniches with different volumes. Colored regions show cell volumes between 2000 ~ 3000, 3000 ~ 4000, 4000 ~ 5000, and >5000 μm3, respectively. The values of cell volumes were presented on each image. Data are shown as mean ± s.d. for all panels, and *P < 0.05, **P < 0.01. Scale bar for all images is 20 μm
Fig. 3
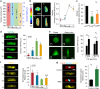
Focal adhesions formation and cell tension in a 3D microniche. a Representative images of vinculin staining for single hMSCs cultured in 3D microniches with different volumes and geometries. b Fluorescent heat maps of ≥20 cells with the same volume (V 3) but different geometries stained for vinculin. c Representative images of myosin IIa in cells of same geometry but different volumes. d Myosin IIa levels (per cell) as a function of cell volume. e Representative images of cells with different geometries but same volume (V 3). f Myosin IIa levels as a function of cell shape (cylinder and triangular prism) or aspect ratio (cubic and cuboid). g Representative images of myosin IIa and F-actin before and after cells with V 3 treated with 50 μM Blebbistatin (Bleb); bar graph shows quantitation of the changes in the level of myosin IIa after treatment with 50 μM Blebbistatin (Bleb). Data are shown as mean ± s.d. for all panels; n = 45–60 cells analyzed for each data point and *P < 0.05, **P < 0.01 (ANOVA using a Tukey post-test). Scale bar for all images is 20 μm
Fig. 4
Nuclear function and transcription factor activity a Nucleus volume and height as a function of cell volume. The volume of nucleus was calculated by fitting the morphology of nucleus to an ellipsoidal shape. Data are given as mean ± s.d. with 12 ≤ n ≤ 15. b Quantitation of nucleus average spatial density (total DAPI intensity per nuclear volume) as a function of cell volume. Highly condensed domains show higher fluorescence intensity. The scale bar is 5 μm. c Quantitation of the changes in the level of nucleus average spatial density for cells with V 3 after treatment with 1 μM cytochalasin D (Cyto D) or 50 μM Blebbistatin (Bleb). d Representative images and quantification of YAP/TAZ localization in hMSCs with different cell volumes but same geometry after 24 h. Scale bar 10 µm. e Representative images and quantification of YAP/TAZ localization in hMSCs with different cell geometries but same volume after 24 h. Scale bar 10 µm. f Representative images of hMSCs stained for HDAC3 on cells with different volumes but same geometry. Histogram shows nuclear HDAC3 levels as a function of cell volume. g Representative images and quantitation of cells with V 3 treated with 50 μM Blebbistatin (Bleb). Data are shown as mean ± s.d. for all panels, n = 50–60 cells analyzed for each data point. *P < 0.05, **P < 0.01 (ANOVA using a Tukey post-test). NS no significant difference
Fig. 5
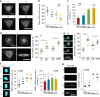
Size and geometry affect mRNA concentration in cells. a Representative images of RhoA mRNA in cells with different volumes. b, c Counts and concentration (divided by cell volume) of RhoA mRNA in cells with different volumes, narrow lines represent the mean within an individual donor; n = 10 cells per donor and condition. d Representative images and total counts of RhoA mRNA in cells with different geometries but same volume (V 3). e Representative images and total counts of Arp2/3 mRNA in cells with different geometries but same volume (V 3). f Representative images, counts, and concentration (divided by nuclear volume) of TEAD1 in cells with different volumes. g Total mRNA in the middle stack of cells with different volumes; we measured total mRNA by quantifying total fluorescence intensity from an mRNA FISH probe that detects polyA tail. Data are shown as mean ± s.d. for all panels; n = 30–35 cells analyzed for each data point. *P < 0.05, **P < 0.01 (ANOVA using a Tukey post-test). NS no significant difference. Scale bar for all images is 20 µm
Fig. 6
Size and geometry affect single hMSC fate. a Alkaline phosphatase (ALP) staining for cells with different V 1 and V 3 volume. The ALP-positive cells were determined by applying an optimal threshold to the image; ALP intensity above the threshold was determined as ALP positive. b Quantification of differentiation after 7 days (ALP) and 10 days (Oil Red O) for cells with different volumes. c Representative images show Oil Red O positive and negative staining for cells with volume (V 3) but different geometries. d Quantification of adipogenic differentiation after 10 days for cells with different geometries. Mean ± s.d., ANOVA one-way analysis followed by Tukey post-hoc test shows significance levels of *P < 0.05, **P < 0.01. NS no significant difference. N ≥ 6 regions of interest (ROI) with a total of 150–200 cells analyzed. Scale bar 20 µm
Similar articles
- Acoustic tweezing cytometry enhances osteogenesis of human mesenchymal stem cells through cytoskeletal contractility and YAP activation.
Xue X, Hong X, Li Z, Deng CX, Fu J. Xue X, et al. Biomaterials. 2017 Jul;134:22-30. doi: 10.1016/j.biomaterials.2017.04.039. Epub 2017 Apr 22. Biomaterials. 2017. PMID: 28453955 Free PMC article. - Multi-lineage differentiation of human mesenchymal stromal cells on the biophysical microenvironment of cell-derived matrix.
Choi DH, Suhaeri M, Hwang MP, Kim IH, Han DK, Park K. Choi DH, et al. Cell Tissue Res. 2014 Sep;357(3):781-92. doi: 10.1007/s00441-014-1898-5. Epub 2014 May 23. Cell Tissue Res. 2014. PMID: 24853672 - Focal adhesion and actin orientation regulated by cellular geometry determine stem cell differentiation via mechanotransduction.
Wang X, Yang Y, Wang Y, Lu C, Hu X, Kawazoe N, Yang Y, Chen G. Wang X, et al. Acta Biomater. 2024 Jul 1;182:81-92. doi: 10.1016/j.actbio.2024.05.017. Epub 2024 May 9. Acta Biomater. 2024. PMID: 38734287 - [Research status of mechanical stimulation of stem cells differentiation in stem cells microenvironment].
Cui S, Zhao W, Yu S, Xing G, Zhao F. Cui S, et al. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2014 Jan;28(1):100-4. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2014. PMID: 24693789 Review. Chinese. - Functional morphometric analysis in cellular behaviors: shape and size matter.
Yu H, Lim KP, Xiong S, Tan LP, Shim W. Yu H, et al. Adv Healthc Mater. 2013 Sep;2(9):1188-97. doi: 10.1002/adhm.201300053. Epub 2013 May 27. Adv Healthc Mater. 2013. PMID: 23713066 Review.
Cited by
- The Roles of YAP/TAZ and the Hippo Pathway in Healthy and Diseased Skin.
Rognoni E, Walko G. Rognoni E, et al. Cells. 2019 May 3;8(5):411. doi: 10.3390/cells8050411. Cells. 2019. PMID: 31058846 Free PMC article. Review. - Implications of Cellular Mechanical Memory in Bioengineering.
Dudaryeva OY, Bernhard S, Tibbitt MW, Labouesse C. Dudaryeva OY, et al. ACS Biomater Sci Eng. 2023 Nov 13;9(11):5985-5998. doi: 10.1021/acsbiomaterials.3c01007. Epub 2023 Oct 5. ACS Biomater Sci Eng. 2023. PMID: 37797187 Free PMC article. Review. - Protease-degradable hydrogels with multifunctional biomimetic peptides for bone tissue engineering.
Oliver-Cervelló L, Martin-Gómez H, Gonzalez-Garcia C, Salmeron-Sanchez M, Ginebra MP, Mas-Moruno C. Oliver-Cervelló L, et al. Front Bioeng Biotechnol. 2023 Jun 1;11:1192436. doi: 10.3389/fbioe.2023.1192436. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 37324414 Free PMC article. - Effect of Functionalization of Texturized Polypropylene Surface by Silanization and HBII-RGD Attachment on Response of Primary Abdominal and Vaginal Fibroblasts.
Quiles MT, Rodríguez-Contreras A, Guillem-Marti J, Punset M, Sánchez-Soto M, López-Cano M, Sabadell J, Velasco J, Armengol M, Manero JM, Arbós MA. Quiles MT, et al. Polymers (Basel). 2024 Feb 29;16(5):667. doi: 10.3390/polym16050667. Polymers (Basel). 2024. PMID: 38475352 Free PMC article. - Osmotic Pressure and Its Biological Implications.
Zheng S, Li Y, Shao Y, Li L, Song F. Zheng S, et al. Int J Mol Sci. 2024 Mar 14;25(6):3310. doi: 10.3390/ijms25063310. Int J Mol Sci. 2024. PMID: 38542282 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources