The emerging role of systems biology for engineering protein production in CHO cells - PubMed (original) (raw)
Review
The emerging role of systems biology for engineering protein production in CHO cells
Chih-Chung Kuo et al. Curr Opin Biotechnol. 2018 Jun.
Abstract
To meet the ever-growing demand for effective, safe, and affordable protein therapeutics, decades of intense efforts have aimed to maximize the quantity and quality of recombinant proteins produced in CHO cells. Bioprocessing innovations and cell engineering efforts have improved product titer; however, uncharacterized cellular processes and gene regulatory mechanisms still hinder cell growth, specific productivity, and protein quality. Herein, we summarize recent advances in systems biology and data-driven approaches aiming to unravel how molecular pathways, cellular processes, and extrinsic factors (e.g. media supplementation) influence recombinant protein production. In particular, as the available omics data for CHO cells continue to grow, predictive models and screens will be increasingly used to unravel the biological drivers of protein production, which can be used with emerging genome editing technologies to rationally engineer cells to further control the quantity, quality and affordability of many biologic drugs.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures
Fig 1. Three waves of different technologies have enabled continued improvement of recombinant protein production in CHO cells
(a) Recombinant protein production has steadily improved over the past few decades thanks to innovations in bioprocessing, targeted genetic manipulation of cells, and systems biology approaches. Together, novel technologies, approaches and discoveries in each field have been of great importance. (b) A comprehensive survey of the CHO bioprocessing literature [60] highlights the historical development of the field in CHO cell research. The first wave-bioprocess development has been driving most of the earlier studies, while the targeted genetic manipulation, omics studies, and modeling efforts have become increasingly important after the mid-2000s with the increased prevalence of genomic resources, genome editing technologies, and development of novel computational models and algorithms.
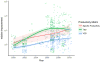
Fig. 2. Published specific productivity, cell density and total product titer has improved steadily over the years
The trend for three major productivity metrics reported by literature from 2000- 2010 [60]. As a result of development in bioprocessing and feeding strategies, the volumetric yield has been greatly improved. The introduction of cell engineering to CHO has further improved the per-cell productivity since the mid-2000s.
Similar articles
- The emerging CHO systems biology era: harnessing the 'omics revolution for biotechnology.
Kildegaard HF, Baycin-Hizal D, Lewis NE, Betenbaugh MJ. Kildegaard HF, et al. Curr Opin Biotechnol. 2013 Dec;24(6):1102-7. doi: 10.1016/j.copbio.2013.02.007. Epub 2013 Mar 20. Curr Opin Biotechnol. 2013. PMID: 23523260 Review. - Optimizing eukaryotic cell hosts for protein production through systems biotechnology and genome-scale modeling.
Gutierrez JM, Lewis NE. Gutierrez JM, et al. Biotechnol J. 2015 Jul;10(7):939-49. doi: 10.1002/biot.201400647. Epub 2015 Jun 23. Biotechnol J. 2015. PMID: 26099571 Review. - The art of CHO cell engineering: A comprehensive retrospect and future perspectives.
Fischer S, Handrick R, Otte K. Fischer S, et al. Biotechnol Adv. 2015 Dec;33(8):1878-96. doi: 10.1016/j.biotechadv.2015.10.015. Epub 2015 Oct 31. Biotechnol Adv. 2015. PMID: 26523782 Review. - CHO-Omics Review: The Impact of Current and Emerging Technologies on Chinese Hamster Ovary Based Bioproduction.
Stolfa G, Smonskey MT, Boniface R, Hachmann AB, Gulde P, Joshi AD, Pierce AP, Jacobia SJ, Campbell A. Stolfa G, et al. Biotechnol J. 2018 Mar;13(3):e1700227. doi: 10.1002/biot.201700227. Epub 2017 Nov 15. Biotechnol J. 2018. PMID: 29072373 Review. - CHO cells in biotechnology for production of recombinant proteins: current state and further potential.
Kim JY, Kim YG, Lee GM. Kim JY, et al. Appl Microbiol Biotechnol. 2012 Feb;93(3):917-30. doi: 10.1007/s00253-011-3758-5. Epub 2011 Dec 9. Appl Microbiol Biotechnol. 2012. PMID: 22159888 Review.
Cited by
- Integrative overview of antibodies against SARS-CoV-2 and their possible applications in COVID-19 prophylaxis and treatment.
Valdez-Cruz NA, García-Hernández E, Espitia C, Cobos-Marín L, Altamirano C, Bando-Campos CG, Cofas-Vargas LF, Coronado-Aceves EW, González-Hernández RA, Hernández-Peralta P, Juárez-López D, Ortega-Portilla PA, Restrepo-Pineda S, Zelada-Cordero P, Trujillo-Roldán MA. Valdez-Cruz NA, et al. Microb Cell Fact. 2021 Apr 22;20(1):88. doi: 10.1186/s12934-021-01576-5. Microb Cell Fact. 2021. PMID: 33888152 Free PMC article. Review. - Gene activation guided by nascent RNA-bound transcription factors.
Liang Y, Xu H, Cheng T, Fu Y, Huang H, Qian W, Wang J, Zhou Y, Qian P, Yin Y, Xu P, Zou W, Chen B. Liang Y, et al. Nat Commun. 2022 Nov 28;13(1):7329. doi: 10.1038/s41467-022-35041-7. Nat Commun. 2022. PMID: 36443367 Free PMC article. - Downscaling screening cultures in a multifunctional bioreactor array-on-a-chip for speeding up optimization of yeast-based lactic acid bioproduction.
Totaro D, Rothbauer M, Steiger MG, Mayr T, Wang HY, Lin YS, Sauer M, Altvater M, Ertl P, Mattanovich D. Totaro D, et al. Biotechnol Bioeng. 2020 Jul;117(7):2046-2057. doi: 10.1002/bit.27338. Epub 2020 Apr 6. Biotechnol Bioeng. 2020. PMID: 32190900 Free PMC article. - Detection of host cell microprotein impurities in antibody drug products.
Tzani I, Castro-Rivadeneyra M, Kelly P, Strasser L, Zhang L, Clynes M, Karger BL, Barron N, Bones J, Clarke C. Tzani I, et al. Nat Commun. 2024 Oct 4;15(1):8605. doi: 10.1038/s41467-024-51870-0. Nat Commun. 2024. PMID: 39366928 Free PMC article. - Proteogenomic Annotation of Chinese Hamsters Reveals Extensive Novel Translation Events and Endogenous Retroviral Elements.
Li S, Cha SW, Heffner K, Hizal DB, Bowen MA, Chaerkady R, Cole RN, Tejwani V, Kaushik P, Henry M, Meleady P, Sharfstein ST, Betenbaugh MJ, Bafna V, Lewis NE. Li S, et al. J Proteome Res. 2019 Jun 7;18(6):2433-2445. doi: 10.1021/acs.jproteome.8b00935. Epub 2019 May 8. J Proteome Res. 2019. PMID: 31020842 Free PMC article.
References
- Lewis NE, Liu X, Li Y, Nagarajan H, Yerganian G, O'Brien E, Bordbar A, Roth AM, Rosenbloom J, Bian C, et al. Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome. Nat Biotechnol. 2013;31:759–765. - PubMed
- Wurm FM, Hacker D. First CHO genome. Nat Biotechnol. 2011;29:718–720. - PubMed
- Kantardjieff A, Jacob NM, Yee JC, Epstein E, Kok YJ, Philp R, Betenbaugh M, Hu WS. Transcriptome and proteome analysis of Chinese hamster ovary cells under low temperature and butyrate treatment. J Biotechnol. 2010;145:143–159. - PubMed
- Rita Costa A, Elisa Rodrigues M, Henriques M, Azeredo J, Oliveira R. Guidelines to cell engineering for monoclonal antibody production. Eur J Pharm Biopharm. 2010;74:127–138. - PubMed
- Nishimiya D. Proteins improving recombinant antibody production in mammalian cells. Appl Microbiol Biotechnol. 2014;98:1031–1042. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources