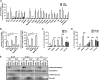A Large Polysaccharide Produced by Helicobacter hepaticus Induces an Anti-inflammatory Gene Signature in Macrophages - PubMed (original) (raw)
A Large Polysaccharide Produced by Helicobacter hepaticus Induces an Anti-inflammatory Gene Signature in Macrophages
Camille Danne et al. Cell Host Microbe. 2017.
Abstract
Interactions between the host and its microbiota are of mutual benefit and promote health. Complex molecular pathways underlie this dialog, but the identity of microbe-derived molecules that mediate the mutualistic state remains elusive. Helicobacter hepaticus is a member of the mouse intestinal microbiota that is tolerated by the host. In the absence of an intact IL-10 signaling, H. hepaticus induces an IL-23-driven inflammatory response in the intestine. Here we investigate the interactions between H. hepaticus and host immune cells that may promote mutualism, and the microbe-derived molecule(s) involved. Our results show that H. hepaticus triggers early IL-10 induction in intestinal macrophages and produces a large soluble polysaccharide that activates a specific MSK/CREB-dependent anti-inflammatory and repair gene signature via the receptor TLR2. These data identify a host-bacterial interaction that promotes mutualistic mechanisms at the intestinal interface. Further understanding of this pathway may provide novel prevention and treatment strategies for inflammatory bowel disease.
Keywords: CREB; Helicobacter hepaticus; MSK1/2; TLR2; anti-inflammatory gene signature; host-microbe interactions; inflammatory bowel disease; macrophage; mutualism; polysaccharide.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
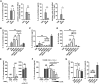
H. hepaticus Induces IL-10 in Gut-Resident Macrophages (A–D) SPF WT mice were infected with Helicobacter hepaticus (Hh) for 3 or 5 days. (A) Experimental design. (B and C) FACS analysis of caecum and colon LPL after 3 days of infection with Hh. (B) Frequency of resident macrophages among total CD45+ cells. (C) Frequency of IL-10high cells among resident macrophages, using an anti-mouse IL-10 antibody or its isotype control. (D) Expression level of Il10, Il6, and Tnf mRNA in caecum or colon tissue after 3 or 5 days of infection with Hh. Each symbol represents an individual mouse (two to three independent experiments). Mann-Whitney test (B and C) or one-way ANOVA and Tukey’s multiple comparisons test (D), p < 0.05. See also Figure S1.
Figure 2
H. hepaticus Produces a Large Soluble Polysaccharide-Inducing IL-10 Production in Macrophages (A–G) M-CSF-differentiated BMDMs were stimulated for 3 hr with different culture fractions of H. hepaticus. (A) mRNA and protein induction of the cytokines IL-10 and IL-6 after stimulation with control medium (TSB), H. hepaticus whole bacteria (Hh), or H. hepaticus_-filtered cultured supernatant (SN_Hh). (B–D) Induction of (B) IL-10, (C) IL-6, and (D) IL-10/IL-6 ratio after 3 hr stimulation with SN_Hh_ treated with enzymes and heat (SN_Hh_t), SN_Hh_t fractionated by size (SN_Hh_t > 30 kDa and SN_Hh_t < 30 kDa), and LPS UP. (E) Induction of IL-10 after stimulation with TSBt or SN_Hh_t, treated with buffer (−) or sodium metaperiodate (+, NaIO4). Stimulation with TSBt ± NaIO4 and LPS ST is used as a positive control for IL-10 production by BMDMs. (F) Induction of IL-10 after stimulation with TSBt, LPS ST, SN_Hh_t, or SN_Hh_t depleted using increasing concentrations of ConA-lectin beads (0.8%, 1.5%, 3%, and 6% v/v). (G) Induction of IL-10 and IL-6 after stimulation with SN_Hh_t crude polysaccharide fraction extracted by a cold-ethanol precipitation method (CP_Hh_t). Data from three independent experiments. Mann-Whitney test, p < 0.05. Mean ± SD. LPS UP, LPS ultrapure from E. coli O111:B4; LPS ST, LPS standard from E. coli O55:B5. See also Figure S2.
Figure 3
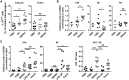
SN_Hh_t Is Sufficient to Induce IL-10 In Vivo (A) Frequency of IL-10high cells among resident macrophages from caecum and colon LPL. SPF WT mice were infected with Hh or orally gavaged with TSBt or SN_Hh_t for 3 days. (B and C) Il10, Il6, and Tnf mRNA expression levels in the peritoneal cell fraction after 2 days (B) or Il10 and Il6 mRNA transcripts after 6 hr (C) challenge. Ligands (TSBt and SN_Hh_t, 200 μL; LPS UP and Pam3, 50 μg) were injected into the intraperitoneal cavity of SPF WT mice. Each symbol represents an individual mouse (two to three independent experiments). Pam3, Pam3CSK4. Mann-Whitney test, p < 0.05. See also Figure S3.
Figure 4
SN_Hh_t Signals through TLR2 and MyD88 to Induce IL-10 (A) Induction of IL-10 production in different knockout C57BL/6 BMDMs stimulated for 3 hr with SN_Hh_t normalized to WT BMDMs (three independent experiments). Two-way ANOVA and Bonferroni’s multiple comparisons test, p < 0.05. Ifnar, interferon-a/b receptor; Mr, mannose receptor; Sra, scavenger receptor A. (B) Il10 mRNA and protein induction in different knockout BMDMs stimulated for 3 hr with SN_Hh_t (one of three independent experiments). One-way ANOVA and Tukey’s multiple comparisons test, p < 0.05. (C) Induction of IL-10 production in WT and _Tlr2_−/− BMDMs stimulated for 3 hr with TSBt, SN_Hh_t, CP_Hh_t (SN_Hh_t crude polysaccharide fraction), LPS UP (TLR4 ligand), or Pam3 (TLR2/1) (one of three independent experiments). Two-way ANOVA and Sidak’s multiple comparisons test, p < 0.05. (D) Induction of IL-10 production in WT BMDMs pre-treated for 2 hr with a blocking antibody specifically inhibiting TLR2 signaling (monoclonal α-mTLR2-IgG) or its isotype control before stimulation for 3 hr with TSBt, SN_Hh_t, Pam2 (TLR2/6), Pam3, or LPS UP (three independent experiments). Two-way ANOVA and Sidak’s multiple comparisons test, p < 0.05. (E) Western blot showing the phosphorylation of ERK1/2 and p38 and the total ERK1/2 and p38 protein amounts in WT and _Tlr2_−/− BMDMs after stimulation with TSBt, SN_Hh_t, Pam3, or LPS UP for 15 min, 30 min, and 3 hr. Mean ± SD. See also Figure S4.
Figure 5
Differential Regulation of Transcription by SN_Hh_t and Pam3 (A–F) Microarray analysis performed on M-CSF BMDMs differentiated from 5 WT mice and stimulated for 3 hr. (A) Principle components analysis (PCA) of transcriptional profiles (empirical Bayes normalized expression values from LIMMA) across three conditions (TSBt, Pam3, and SN_Hh_t, n = 5/condition, 16,692 probes). (B) Hierarchical clustering heatmap (Manhattan distance with Ward clustering of empirical Bayes normalized expression values) of the union of differentially expressed probes between any condition-condition contrast (i.e., TSBt versus Pam3, TSBt versus SN_Hh_t, or Pam3 versus SN_Hh_t) (n = 172 probes, n = 146 genes). Probes were assigned to clusters using k-means clustering (k = 4) and cluster assignments are annotated as colors to the left of the heatmap. The number of probes assigned to each cluster were: cluster 1 = 75, cluster 2 = 10, cluster 3 = 68, and cluster 4 = 19. (C) Scatterplot displaying the relationship between log2 (fold changes) for each gene shown in (B) obtained when comparing either Pam3 (x axis) or SN_Hh_t (y axis) with the TSBt control condition. Colors represent the cluster assignments. (D and E) Transcription factor (TF) motif enrichment analysis among genes assigned to (D) cluster 4 (i.e., specifically induced by SN_Hh_t) and (D) cluster 1 (i.e., specifically induced by Pam3). Motif names correspond to Transfac motif identifiers derived from the Molecular Signatures Database (MSigDB). Motifs significantly overrepresented at an adjusted p < 0.05 (permutation test) are shown. (F) mRNA levels of CREB (Il10, Fosb, Egr3, Rcan1) and NF-κB (Il6, Ccl5, Cd40, Icam1) target genes were validated by qRT-PCR.
Figure 6
SN_Hh_t Stimulates CREB Phosphorylation and Induction of an Anti-inflammatory and Repair Gene Signature in Macrophages (A) Western blot showing the phosphorylation of CREB1 S133 and RelA S536 and the total CREB1 and RelA protein amounts in WT and _Tlr2_−/− BMDMs after 30 min stimulation with TSBt, SN_Hh_t, Pam3, or LPS UP. β-actin used as a loading control. (B) mRNA levels of Il10, Fosb, and Egr3 genes in BMDMs from conditional vav-cre CREBWT and CREBS133A KI mice after 1 hr stimulation with TSBt, Hh, SN_Hh_t, or Pam3. (C) Induction of IL-10, IL-6, and TNFα; IL-10/IL-6; and IL-10/TNFα protein ratios in BMDMs from conditional vav-cre CREBWT and CREBS133A KI mice after 10 hr stimulation with TSBt, Hh, SN_Hh_t, or Pam3. One of three independent experiments. Two-way ANOVA and Sidak’s and/or Tukey’s multiple comparisons tests, p < 0.05. (D) mRNA levels of Il10, Fosb, Egr3, Rcan1, Il6, Tnf, and Cd40 genes in caecum total tissue from WT mice gavaged with TSBt or SN_Hh_t for 2 days. Each symbol represents an individual mouse (two independent experiments). Mann-Whitney test, p < 0.05. Mean ± SD. See also Figure S5.
Figure 7
MSK1/2 Is Essential to the Anti-inflammatory Properties of SN_Hh_t (A) Western blot showing the phosphorylation of CREB1 S133 and MSK1 T581 and the total CREB1 and MSK1 protein amounts in WT and _Msk1/2_−/− BMDMs after 30 min stimulation with TSBt, SN_Hh_t, Pam3, or LPS UP. (B) mRNA levels of Il10, Fosb, and Egr3 genes in BMDMs from WT or _Msk1/2_−/− mice after 1 hr stimulation. (C) Induction of IL-10, IL-6, and TNFα; IL-10/IL-6; and IL-10/TNFα protein ratios in BMDMs from WT or _Msk1/2_−/− mice after 10 hr stimulation. One of three independent experiments. Two-way ANOVA and Sidak’s and/or Tukey’s multiple comparisons tests, p < 0.05. Mean ± SD. See also Figure S6.
Comment in
- Sweet! Helicobacter Sugar Calms Intestinal Macrophages.
Bain CC, Kullberg MC. Bain CC, et al. Cell Host Microbe. 2017 Dec 13;22(6):719-721. doi: 10.1016/j.chom.2017.11.014. Cell Host Microbe. 2017. PMID: 29241034 - Helicobacter hepaticus polysaccharide induces an anti-inflammatory response in intestinal macrophages.
Danne C, Powrie F. Danne C, et al. Microb Cell. 2018 Mar 22;5(4):208-211. doi: 10.15698/mic2018.04.626. Microb Cell. 2018. PMID: 29611556 Free PMC article.
References
- Ananieva O., Darragh J., Johansen C., Carr J.M., McIlrath J., Park J.M., Wingate A., Monk C.E., Toth R., Santos S.G. The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nat. Immunol. 2008;9:1028–1036. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases