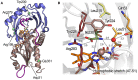Competitive Inhibitors Unveil Structure/Function Relationships in Human D-Amino Acid Oxidase - PubMed (original) (raw)
Review
Competitive Inhibitors Unveil Structure/Function Relationships in Human D-Amino Acid Oxidase
Gianluca Molla. Front Mol Biosci. 2017.
Abstract
D-amino acid oxidase (DAAO) catalyzes the oxidative deamination of several neutral D-amino acids and is the enzyme mainly responsible (together with serine racemase) for degrading D-serine (D-Ser) in the central nervous system of mammals. This D-amino acid, which binds the coagonist site of the N-methyl-D-aspartate receptor, is thus a key neuromodulator of glutamatergic neurotransmission. Altered D-Ser metabolism results in several pathological conditions (e.g., amylotrophic lateral sclerosis or schizophrenia, SZ) for which effective "broad spectrum" pharmaceutical drugs are not yet available. In particular, the correlation between reduced D-Ser concentration and SZ led to a renaissance of biochemical interest in human DAAO (hDAAO). In the last 10 years, public and corporate research laboratories undertook huge efforts to study the structural, enzymatic, and physiological properties of the human flavoenzyme and to identify novel effective inhibitors which, acting as pharmaceutical drugs, could decrease hDAAO activity, thus restoring the physiological concentration of D-Ser. Although, none of the identified hDAAO inhibitors has reached the market yet, from a biochemical point of view, these compounds turned out to be invaluable for gaining a detailed understanding of the structure/function relationships at the molecular level in the mammalian DAAO, in particular of the interaction between ligand and the enzyme. This detailed knowledge, together with several recent studies concerning the interaction of the human enzyme with other protein regulative partners, its subcellular localization, and in vivo degradation, contributed to gaining comprehensive knowledge of the structure, function, and physiopathological role of this important human enzyme.
Keywords: D-amino acid oxidase; D-serine; drug design; flavoprotein; inhibitor; neurotransmission; schizophrenia; structure.
Figures
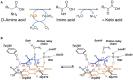
Figure 1
Reaction catalyzed by human D-amino acid oxidase. (A) Scheme of the reaction. R1 represents the side chain of the substrate. (B) Schematic representation of the hydride transfer mechanisms. The D-amino acid is bound in its zwitterionic form. Substrate is colored in blue. H-bonds are represented as dotted lines. Arrows, indicating relocation of electrons during the reaction, are represented in gray. Wat: putative active-site water molecule. R1: side chain of the substrate. The first H acceptor of the proposed proton relay chain (at pH < 8) has not yet been identified: it could be an active-site water molecule (R2 = H) or the hydroxyl of a protein residues (R2 = protein).
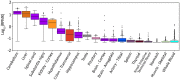
Figure 2
Expression pattern of DAO gene in different tissues. Box plot of expression levels of DAO gene in different human tissues. Values are shown in RPKM (reads per kilobase of transcript per million mapped reads), calculated from a gene model with isoforms collapsed to a single gene. No other normalization steps have been applied. Box plots are shown as median and 25th and 75th percentiles; points are displayed as outliers if they are above or below 1.5 times the interquartile range. The data presented in this plot were obtained from the Genotype-Tissue Expression (GTEx) Project Portal, dbGaP accession number phs000424.v6.p1 on 09/04/2017 (
https://www.gtexportal.org/home/gene/DAO
).
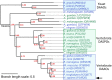
Figure 3
Schematic phylogenetic tree of DAAOs and DASPOs from selected model species. Yeast DAAOs represent the outgroup allowing the phylogenetic tree rooting. Selected sequences were aligned using Clustal Omega Server. Alignment was used to calculate a maximum-likelihood phylogenetic tree using PhyML 3.0 server (Guindon et al., 2010). hDAAO is indicated by an arrow. Sequences were identified by their UniProtKB accession number. DAAO sequences are in blue while DASPO sequences are in green. Supporting values for each node are represented in red. Branch length scale units represent the nucleotide substitutions per site.
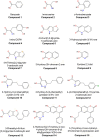
Figure 4
Chemical structures of human D-amino acid oxidase ligands reported in Table 1.
Figure 5
Three-dimensional structure of hDAAO. (A) Schematic view of the spatial arrangement of protein domains in hDAAO (PDB code 2e49) (Kawazoe et al., 2007b). The FAD binding domain is in dark salmon color and the substrate binding domain is in blue. Regions showing the highest conformational diversity are in purple; α-helices, β-strands, and coils are drawn in cartoon representation. (B) Detail of the active site. The substrate D-Ser was modeled based on the structure of the enzyme in complex with imino-serine. Residues are colored according to their ConSurf score from darker (most conserved) to lighter (less conserved) color. Gly313 αC is represented as a sphere. The “putative” active-site water molecule is in red (see text). The FAD cofactor (orange) and the ligand (green) are shown in stick form. The H-bonds are represented by dotted lines while the distance between the α-H of the substrate and the N(5) of FAD is depicted with a continuous line.
Figure 6
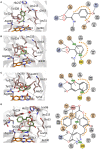
Mode of binding of hDAAO ligands. Human DAAO in complex with (A) imino-DOPA, PDB code 2E82; (B) o-Aminobenzoate, PDB code 2E4A; (C) 3-hydroxy-2H-chromen-2-one, PDB code 3ZNP; (D) 3-(7-hydroxy-2-oxo-4-phenyl-2H-chromen-6-yl)propanoic acid, PDB code 4QFD. Left: detail of the active site. The ligand is in green and the FAD in orange. Dotted lines represent H-bonds and thick brown lines represent π-π interactions. Right: schematic view of ligand-protein interactions. Dotted lines represent H-bonds and π-π interactions. Pale orange dots represent hydrophobic contacts.
Figure 7
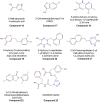
Chemical structures of human D-amino acid oxidase inhibitors reported in Table 3.
Similar articles
- Biochemical Properties of Human D-amino Acid Oxidase Variants and Their Potential Significance in Pathologies.
Sacchi S, Cappelletti P, Murtas G. Sacchi S, et al. Front Mol Biosci. 2018 Jun 12;5:55. doi: 10.3389/fmolb.2018.00055. eCollection 2018. Front Mol Biosci. 2018. PMID: 29946548 Free PMC article. Review. - Biochemical Properties of Human D-Amino Acid Oxidase.
Murtas G, Sacchi S, Valentino M, Pollegioni L. Murtas G, et al. Front Mol Biosci. 2017 Dec 15;4:88. doi: 10.3389/fmolb.2017.00088. eCollection 2017. Front Mol Biosci. 2017. PMID: 29326945 Free PMC article. - Structure-function relationships in human d-amino acid oxidase variants corresponding to known SNPs.
Cappelletti P, Piubelli L, Murtas G, Caldinelli L, Valentino M, Molla G, Pollegioni L, Sacchi S. Cappelletti P, et al. Biochim Biophys Acta. 2015 Sep;1854(9):1150-9. doi: 10.1016/j.bbapap.2015.02.005. Epub 2015 Feb 17. Biochim Biophys Acta. 2015. PMID: 25701391 - Human D-Amino Acid Oxidase: Structure, Function, and Regulation.
Pollegioni L, Sacchi S, Murtas G. Pollegioni L, et al. Front Mol Biosci. 2018 Nov 28;5:107. doi: 10.3389/fmolb.2018.00107. eCollection 2018. Front Mol Biosci. 2018. PMID: 30547037 Free PMC article. Review. - Yin and Yang in Post-Translational Modifications of Human D-Amino Acid Oxidase.
Sacchi S, Rabattoni V, Miceli M, Pollegioni L. Sacchi S, et al. Front Mol Biosci. 2021 May 10;8:684934. doi: 10.3389/fmolb.2021.684934. eCollection 2021. Front Mol Biosci. 2021. PMID: 34041270 Free PMC article.
Cited by
- Canonical and Non-Canonical Antipsychotics' Dopamine-Related Mechanisms of Present and Next Generation Molecules: A Systematic Review on Translational Highlights for Treatment Response and Treatment-Resistant Schizophrenia.
de Bartolomeis A, Ciccarelli M, De Simone G, Mazza B, Barone A, Vellucci L. de Bartolomeis A, et al. Int J Mol Sci. 2023 Mar 21;24(6):5945. doi: 10.3390/ijms24065945. Int J Mol Sci. 2023. PMID: 36983018 Free PMC article. Review. - d-Amino Acids Are Exuded by Arabidopsis thaliana Roots to the Rhizosphere.
Hener C, Hummel S, Suarez J, Stahl M, Kolukisaoglu Ü. Hener C, et al. Int J Mol Sci. 2018 Apr 7;19(4):1109. doi: 10.3390/ijms19041109. Int J Mol Sci. 2018. PMID: 29642439 Free PMC article. - Comparative Pro-cognitive and Neurochemical Profiles of Glycine Modulatory Site Agonists and Glycine Reuptake Inhibitors in the Rat: Potential Relevance to Cognitive Dysfunction and Its Management.
Fone KCF, Watson DJG, Billiras RI, Sicard DI, Dekeyne A, Rivet JM, Gobert A, Millan MJ. Fone KCF, et al. Mol Neurobiol. 2020 May;57(5):2144-2166. doi: 10.1007/s12035-020-01875-9. Epub 2020 Jan 20. Mol Neurobiol. 2020. PMID: 31960362 Free PMC article. - Structural basis for potent inhibition of d-amino acid oxidase by thiophene carboxylic acids.
Kato Y, Hin N, Maita N, Thomas AG, Kurosawa S, Rojas C, Yorita K, Slusher BS, Fukui K, Tsukamoto T. Kato Y, et al. Eur J Med Chem. 2018 Nov 5;159:23-34. doi: 10.1016/j.ejmech.2018.09.040. Epub 2018 Sep 18. Eur J Med Chem. 2018. PMID: 30265959 Free PMC article. - Directly and Indirectly Targeting the Glycine Modulatory Site to Modulate NMDA Receptor Function to Address Unmet Medical Needs of Patients With Schizophrenia.
Pei JC, Luo DZ, Gau SS, Chang CY, Lai WS. Pei JC, et al. Front Psychiatry. 2021 Oct 1;12:742058. doi: 10.3389/fpsyt.2021.742058. eCollection 2021. Front Psychiatry. 2021. PMID: 34658976 Free PMC article. Review.
References
- Adage T., Trillat A. C., Quattropani A., Perrin D., Cavarec L., Shaw J., et al. . (2008). In vitro and in vivo pharmacological profile of AS057278, a selective D-amino acid oxidase inhibitor with potential anti-psychotic properties. Eur. Neuropsychopharmacol. 18, 200–214. 10.1016/j.euroneuro.2007.06.006 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources