Mechanistic Investigations of PoyD, a Radical S-Adenosyl-l-methionine Enzyme Catalyzing Iterative and Directional Epimerizations in Polytheonamide A Biosynthesis - PubMed (original) (raw)
Mechanistic Investigations of PoyD, a Radical S-Adenosyl-l-methionine Enzyme Catalyzing Iterative and Directional Epimerizations in Polytheonamide A Biosynthesis
Aubérie Parent et al. J Am Chem Soc. 2018.
Abstract
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a growing family of bioactive peptides. Among RiPPs, the bacterial toxin polytheonamide A is characterized by a unique set of post-translational modifications catalyzed by novel radical S-adenosyl-l-methionine (SAM) enzymes. Here we show that the radical SAM enzyme PoyD catalyzes in vitro polytheonamide epimerization in a C-to-N directional manner. By combining mutagenesis experiments with labeling studies and investigating the enzyme substrate promiscuity, we deciphered in detail the mechanism of PoyD. We notably identified a critical cysteine residue as a likely key H atom donor and demonstrated that PoyD belongs to a distinct family of radical SAM peptidyl epimerases. In addition, our study shows that the core peptide directly influences the epimerization pattern allowing for production of peptides with unnatural epimerization patterns.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Figure 1
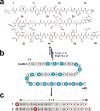
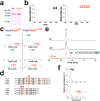
Structure of Polytheonamide A and peptide substrates designed to investigate PoyD mechanism. (a) Structure of polytheonamide A. Numbers indicate amino acid residues location. Methyl groups labeled in blue are inserted by the radial SAM enzyme PoyC, while methyl groups labeled in purple have been proposed to be inserted by the radical SAM enzyme PoyB. Red labels are D-amino acid residues formed by the radical SAM enzyme PoyD. (b) Sequence of PoyA, the peptide precursor of polytheonamide A. Circles filled in blue indicate the amino acid residues epimerized in mature polytheonamide A. The enzymes responsible for post-translational modifications of PoyA are indicated next to the arrows. (c) Sequence of peptides 1 and 2 used as substrates. Circles filled in gray indicate amino acid residues from the leader sequence while circles filled in white indicate amino acids from the core sequence. White circles with a red line are residues epimerized in polytheonamide A. Circles filled in red indicate amino acid residues introduced in the sequence for analytical purpose. Numbers are relative to PoyA sequence with positive numbers for the core peptide and negative numbers for the leader-peptide sequence.
Figure 2
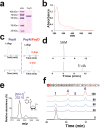
In vitro characterization of PoyD. (a) Gel electrophoresis analysis of purified PoyD expressed in E. coli. MW: Molecular weight markers. (b) UV–visible spectrum of as-purified (dotted line) and anaerobically reconstituted PoyD (plain line). (c) HPLC analysis of the amino acid content of PoyA after its in vivo expression in E. coli alone or in the presence of PoyD (left and right panels respectively). Amino acids were analyzed after acid hydrolysis and derivatization with _N_-α-(2,4-dinitro-5-fluorophenyl)-
l
-valinamide (
l
-FDVA) and their retention times compared with authentic standards. (d) HPLC analysis of SAM incubated with PoyD. Traces indicate incubation under anaerobic conditions in the presence of sodium dithionite at t = 0 and after 90 min. As shown, during incubation _S_-adenosyl-
l
-methionine (SAM) is cleaved in 5′-deoxyadenosine (5′-dA). (e) LC–MS analysis of 5′-deoxyadenosine (5′-dA) produced by PoyD. (f) Activity of PoyD toward peptide 1. HPLC Analyses were performed at t = 0, 30, 60, 90, and 120 min (lower to upper traces respectively). See
Supporting Information
for experimental conditions.
Figure 3
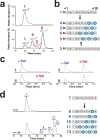
Characterization of the products formed in vitro by PoyD. (a) LC–MS/MS analysis of peptide 1 incubated with PoyD. Upper trace peptide 1. Lower trace, peptide 1 incubated with PoyD for 2 h, in deuterated buffer. See
Supporting Information
for experimental conditions. Numbers refer to the corresponding peptides. (b) Sequence of the different products formed by PoyD in vitro after incubation with peptide 1 or peptide 2. See Figure 1 for the amino acid residues corresponding to R and R′ and
Supplementary Figures S3–4 and S6 and Supplementary Tables S1–S6
for complete peptide assignment. (c) LC–MS/MS analysis of the
l
-/
d
-Val content in peptide 1 and the products: peptides 4, 5, and 6 obtained after incubation with PoyD. Amino acids were analyzed after hydrolysis and derivatization by
l
-FDVA and detected by LC–MS after ion current extraction in MS/MS experiments using the transition 398 > 352 for Val-FDVA derivatives. Numbers indicate peptides analyzed. (d) LC–MS/MS analysis of peptide 7 incubated with PoyD. Analysis was performed at t = 0 and after 2 h incubation, upper and lower traces, respectively. Sequences of the peptides produced are indicated. See
Supplementary Figures S8–S13 and Supplementary Tables S7–S12
for full peptide assignment. Numbers refer to the corresponding peptides.
Figure 4
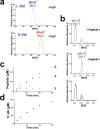
Mechanistic and kinetic analysis of the reaction catalyzed by PoyD. (a) MS/MS spectra of
l
-Val (upper traces) and
d
-Val (lower traces) obtained after incubation of PoyD with peptide 1 in deuterated buffer. Peptide products were purified and after acid hydrolysis, amino acids were derivatized with
l
-FDVA and analyzed by LC–MS/MS. See
Supporting Information
for experimental conditions. (b) MS spectra of 5′-dA produced by PoyD. PoyD was incubated in deuterated buffer under anaerobic and reducing conditions in the presence (upper trace) or in the absence of peptide 1 (middle trace). The lower trace shows MS spectra of 5′-dA produced over time (from t = 0 to t = 240 min) in the presence of peptide 1 in deuterated buffer. Production of epimerized peptides (c) and 5′-dA (d) by PoyD. PoyD was incubated in the presence of peptide 1 under anaerobic conditions with sodium dithionite and SAM. Numbers refer to the corresponding peptides formed.
Figure 5
Identification of a potential H atom donor in PoyD. (a) Gel electrophoresis analysis of the A3 and C372A mutants expressed in E. coli. (b) UV–visible spectrum of the A3 and C372A mutants before (dotted line) and after anaerobic reconstitution (plain line). (c) HPLC analysis of the amino acid content of PoyA after its in vivo expression in E. coli with the A3 or the C372A mutants (left and right panels, respectively). Amino acids were analyzed after acid hydrolysis and derivatization with
l
-FDVA and their retention times compared with authentic standards. See
Supporting Information
for experimental conditions. (d) Sequence alignment between PoyD and other proteusin epimerases OspD, AvpD and PlpD. Strictly conserved residues are highlighted in gray or red (cysteine residues). Numbers refers to amino acid residues location in the respective sequences. (e) HPLC analysis of peptide 1 incubated in the presence of the A3 or C372A mutant. Upper trace: HPLC analysis of peptide 1 at t = 0. Middle trace: HPLC analysis of peptide 1 after 120 min incubation with the A3 mutant. Lower trace: HPLC analysis of peptide 1 after 120 min incubation with the C372A mutant. The sequence of the product formed by the C372A mutant is indicated. See
Supplementary Figure S15 and Supplementary Table S13
for full assignment. (f) LC–MS/MS analysis of the
l
-/
d
-Val content of peptides 1 and 13. Upper trace corresponds to peptide 1 and lower trace to peptide 13 produced by the C372A mutant. LC MS/MS experiments were performed using the transition 398 > 352.
Figure 6
(a) HPLC analysis of peptide 1 after incubation with the C372A mutant and wild-type PoyD. Peptide 1 was incubated with the C372A mutant and analyzed at t = 0 (upper blue trace) and t = 120 (red trace). After purification, peptide 13 was incubated with PoyD and analyzed by HPLC at t = 0 (green trace) and t = 120 min (purple lower trace). Numbers refer to the corresponding peptides. (b) Consumption of peptides 1 and 13 during incubation with PoyD. (c) Production of epimerized peptides by PoyD. PoyD was incubated in the presence of peptide 1 under anaerobic conditions with sodium dithionite and SAM. Numbers refer to the corresponding peptides formed.
Figure 7
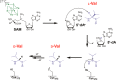
Proposed mechanism for the radical SAM peptide epimerase PoyD. After the reducing SAM cleavage, PoyD generates a 5′-dA•, which abstracts the amino acid Cα H atom. A carbon-centered radical is formed and quenched by the thiolate H atom of Cys-372 leading to the formation of a D-amino acid residue. Reduction of the thiyl radical is likely assisted by other amino acid residues from PoyD similarly to ribonucleotide reductase or spore photoproduct lyase for the next catalytic cycle.
Similar articles
- Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Kubiak X, Polsinelli I, Chavas LMG, Fyfe CD, Guillot A, Fradale L, Brewee C, Grimaldi S, Gerbaud G, Thureau A, Legrand P, Berteau O, Benjdia A. Kubiak X, et al. Nat Chem Biol. 2024 Mar;20(3):382-391. doi: 10.1038/s41589-023-01493-1. Epub 2023 Dec 29. Nat Chem Biol. 2024. PMID: 38158457 - The B12-Radical SAM Enzyme PoyC Catalyzes Valine Cβ-Methylation during Polytheonamide Biosynthesis.
Parent A, Guillot A, Benjdia A, Chartier G, Leprince J, Berteau O. Parent A, et al. J Am Chem Soc. 2016 Dec 7;138(48):15515-15518. doi: 10.1021/jacs.6b06697. Epub 2016 Nov 29. J Am Chem Soc. 2016. PMID: 27934015 Free PMC article. - Direct Detection of the α-Carbon Radical Intermediate Formed by OspD: Mechanistic Insights into Radical _S_-Adenosyl-l-methionine Peptide Epimerization.
Walls WG, Vagstad AL, Delridge T, Piel J, Broderick WE, Broderick JB. Walls WG, et al. J Am Chem Soc. 2024 Feb 28;146(8):5550-5559. doi: 10.1021/jacs.3c13829. Epub 2024 Feb 16. J Am Chem Soc. 2024. PMID: 38364824 Free PMC article. - Radical SAM Enzymes in the Biosynthesis of Ribosomally Synthesized and Post-translationally Modified Peptides (RiPPs).
Benjdia A, Balty C, Berteau O. Benjdia A, et al. Front Chem. 2017 Nov 8;5:87. doi: 10.3389/fchem.2017.00087. eCollection 2017. Front Chem. 2017. PMID: 29167789 Free PMC article. Review. - Peptide Epimerization Machineries Found in Microorganisms.
Ogasawara Y, Dairi T. Ogasawara Y, et al. Front Microbiol. 2018 Feb 6;9:156. doi: 10.3389/fmicb.2018.00156. eCollection 2018. Front Microbiol. 2018. PMID: 29467749 Free PMC article. Review.
Cited by
- Biocatalytic hydrogen atom transfer: an invigorating approach to free-radical reactions.
Nakano Y, Biegasiewicz KF, Hyster TK. Nakano Y, et al. Curr Opin Chem Biol. 2019 Apr;49:16-24. doi: 10.1016/j.cbpa.2018.09.001. Epub 2018 Sep 27. Curr Opin Chem Biol. 2019. PMID: 30269010 Free PMC article. Review. - Identification of the Early Steps in Herbicidin Biosynthesis Reveals an Atypical Mechanism of _C_-Glycosylation.
Chen Z, Sato S, Geng Y, Zhang J, Liu HW. Chen Z, et al. J Am Chem Soc. 2022 Aug 31;144(34):15653-15661. doi: 10.1021/jacs.2c05728. Epub 2022 Aug 18. J Am Chem Soc. 2022. PMID: 35981300 Free PMC article. - New developments in RiPP discovery, enzymology and engineering.
Montalbán-López M, Scott TA, Ramesh S, Rahman IR, van Heel AJ, Viel JH, Bandarian V, Dittmann E, Genilloud O, Goto Y, Grande Burgos MJ, Hill C, Kim S, Koehnke J, Latham JA, Link AJ, Martínez B, Nair SK, Nicolet Y, Rebuffat S, Sahl HG, Sareen D, Schmidt EW, Schmitt L, Severinov K, Süssmuth RD, Truman AW, Wang H, Weng JK, van Wezel GP, Zhang Q, Zhong J, Piel J, Mitchell DA, Kuipers OP, van der Donk WA. Montalbán-López M, et al. Nat Prod Rep. 2021 Jan 1;38(1):130-239. doi: 10.1039/d0np00027b. Epub 2020 Sep 16. Nat Prod Rep. 2021. PMID: 32935693 Free PMC article. Review. - Core-dependent post-translational modifications guide the biosynthesis of a new class of hypermodified peptides.
Pei ZF, Zhu L, Nair SK. Pei ZF, et al. Nat Commun. 2023 Nov 25;14(1):7734. doi: 10.1038/s41467-023-43604-5. Nat Commun. 2023. PMID: 38007494 Free PMC article. - New Role for Radical SAM Enzymes in the Biosynthesis of Thio(seleno)oxazole RiPP Natural Products.
Lewis JK, Jochimsen AS, Lefave SJ, Young AP, Kincannon WM, Roberts AG, Kieber-Emmons MT, Bandarian V. Lewis JK, et al. Biochemistry. 2021 Nov 16;60(45):3347-3361. doi: 10.1021/acs.biochem.1c00469. Epub 2021 Nov 3. Biochemistry. 2021. PMID: 34730336 Free PMC article.
References
- Arnison P. G.; Bibb M. J.; Bierbaum G.; Bowers A. A.; Bugni T. S.; Bulaj G.; Camarero J. A.; Campopiano D. J.; Challis G. L.; Clardy J.; Cotter P. D.; Craik D. J.; Dawson M.; Dittmann E.; Donadio S.; Dorrestein P. C.; Entian K. D.; Fischbach M. A.; Garavelli J. S.; Goransson U.; Gruber C. W.; Haft D. H.; Hemscheidt T. K.; Hertweck C.; Hill C.; Horswill A. R.; Jaspars M.; Kelly W. L.; Klinman J. P.; Kuipers O. P.; Link A. J.; Liu W.; Marahiel M. A.; Mitchell D. A.; Moll G. N.; Moore B. S.; Muller R.; Nair S. K.; Nes I. F.; Norris G. E.; Olivera B. M.; Onaka H.; Patchett M. L.; Piel J.; Reaney M. J.; Rebuffat S.; Ross R. P.; Sahl H. G.; Schmidt E. W.; Selsted M. E.; Severinov K.; Shen B.; Sivonen K.; Smith L.; Stein T.; Sussmuth R. D.; Tagg J. R.; Tang G. L.; Truman A. W.; Vederas J. C.; Walsh C. T.; Walton J. D.; Wenzel S. C.; Willey J. M.; van der Donk W. A. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–60. 10.1039/C2NP20085F. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources