Bidirectional transcription initiation marks accessible chromatin and is not specific to enhancers - PubMed (original) (raw)
Bidirectional transcription initiation marks accessible chromatin and is not specific to enhancers
Robert S Young et al. Genome Biol. 2017.
Abstract
Background: Enhancers are modular regulatory elements that are central to the spatial and temporal regulation of gene expression. Bidirectional transcription initiating at enhancers has been proposed to mark active enhancers and as such has been utilized to experimentally identify active enhancers de novo.
Results: Here, we show that bidirectional transcription initiation is a pervasive feature of accessible chromatin, including at enhancers, promoters, and other DNase hypersensitive regions not marked with canonical histone modification profiles. Transcription is less predictive for enhancer activity than epigenetic modifications such as H3K4me1 or the accessibility of DNA when measured both in enhancer assays and at endogenous loci. The stability of enhancer initiated transcripts does not influence measures of enhancer activity and we cannot detect evidence of purifying selection on the resulting enhancer RNAs within the human population.
Conclusions: Our results indicate that bidirectional transcription initiation from accessible chromatin is not sufficient for, nor specific to, enhancer activity. Transcription initiating at enhancers may be a frequent by-product of promiscuous RNA polymerase initiation at accessible chromatin and is unlikely to generally play a functional role in enhancer activity.
Keywords: Cap analysis of gene expression; Chromatin modifications; DNase hypersensitivity; Enhancer; Gene regulation; Transcription.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Fig. 1
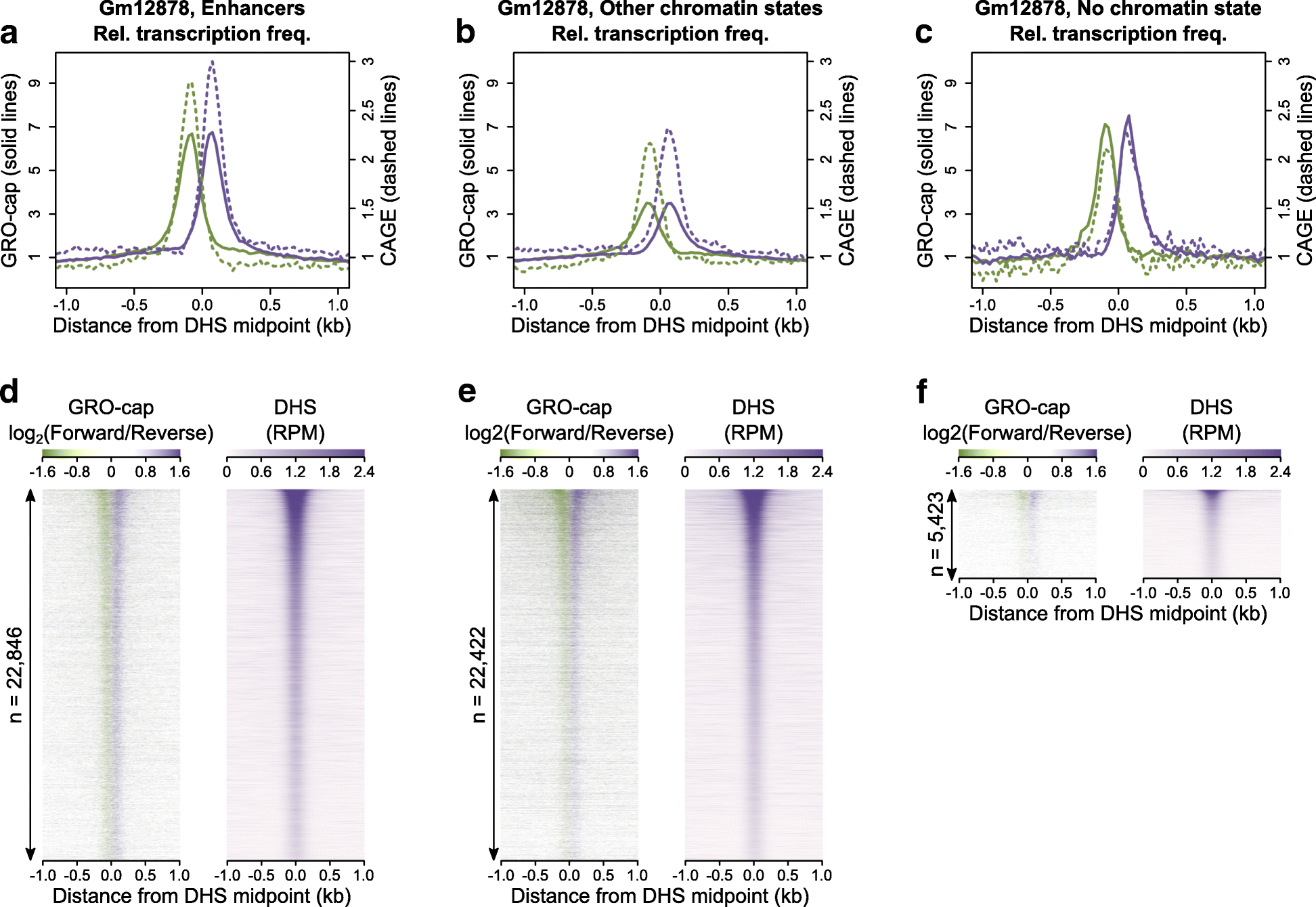
Bidirectional transcription initiates around DHSs but is not a specific mark of active enhancers. a–c The fold-change in transcription frequency (fraction of loci with evidence of transcription initiation) for sites with transcription initiation signal in 25-bp consecutive windows around DHS midpoints (x = 0) relative to the mean transcription frequency in the flanking regions: 500 to 1000 bp from the DHS midpoint. Total bidirectional transcription initiation across DHSs in Gm12878 cells as measured by GRO-cap is shown by the solid lines while stable bidirectional transcription initiation as measured by CAGE is shown by the dashed lines. Purple lines consider transcription initiation from the forward strand and green lines show transcription initiation from the reverse strand. In all panels, only DHSs that do not overlap annotated promoters were included. d–f Heatmaps of GRO-cap signal as measured by log2(Forward/Reverse) RPM and DNase hypersensitivity as measured by RPM around DHS midpoints for the same chromatin state annotations described in a–c. Rows are ranked by the DNase hypersensitivity signal (RPM). The height of each heatmap corresponds to the total number of DHSs which generated the plot as shown on the y-axis so that shading density is directly comparable between plots
Fig. 2
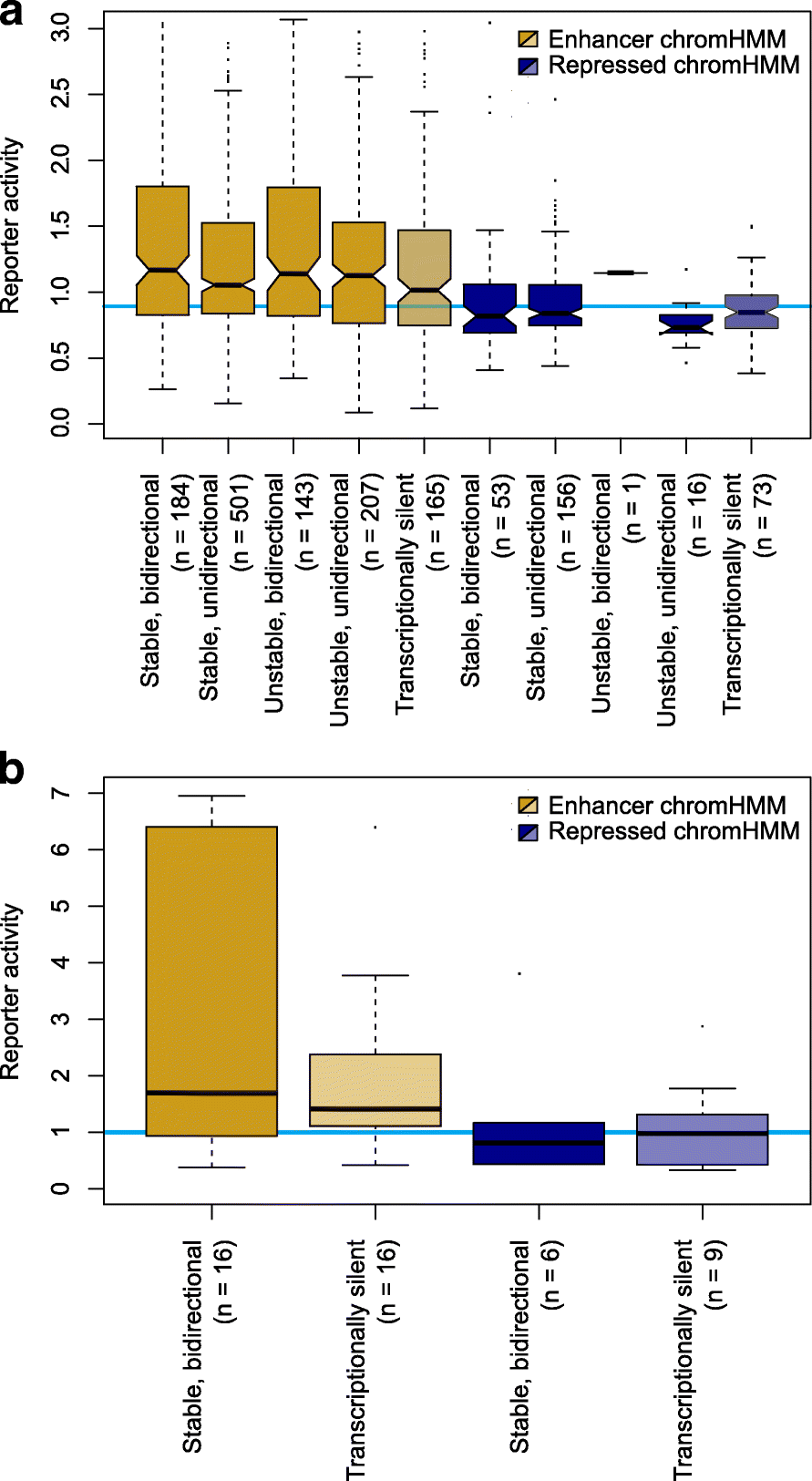
Chromatin-defined enhancer marks rather than transcription are indicative of enhancer activity. a Reporter activities for chromatin mark-defined enhancer and repressed regions in K562 cells with stable and unstable bidirectional and unidirectional transcription initiation, and those with no evidence for transcription. The horizontal blue line indicates the median reporter activity for all scrambled control sequence assays in K562 cells. b As for a, in HepG2 cells but only considering transcribed regions to be those with stable, bidirectional transcription initiation. The blue line indicates the reporter activity for the transfected empty vector
Fig. 3
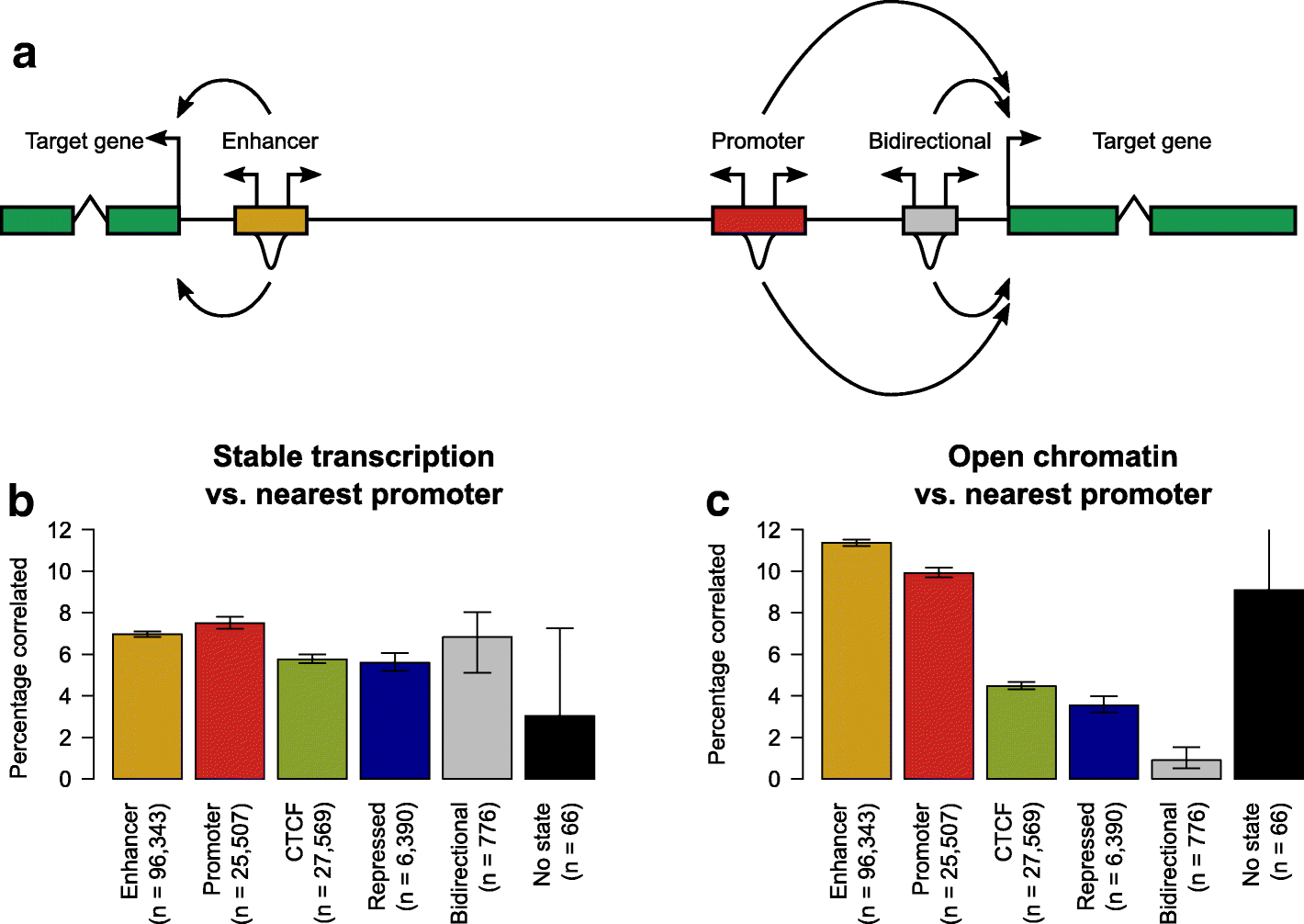
Stable transcription is not indicative of enhancer activity. a As shown by the curved arrows, the putative target of each chromatin state locus and bidirectionally transcribed-defined enhancer is defined as the nearest annotated gene (shown in the green boxes). The activity of each locus as measured by either the level of transcription initiation (the bidirectional arrows above each regulatory region) or the strength of the DHS signal (the peaks below each regulatory region) is then correlated with transcription initiation at the putative target gene promoter. b The percentage of chromatin state loci and bidirectionally transcribed-defined enhancers whose measure of stable transcription initiation is significantly correlated with transcription initiation from the nearest annotated gene promoter. The error bars represent the 95% confidence interval from 1000 samplings of the data with replacement, while the numbers below each bar denote the number of loci tested for a significant correlation. c As for b, but the correlations being considered are between the level of DHS signal of chromatin state loci and bidirectionally transcribed-defined enhancers and transcription initiation from the nearest annotated gene promoter
Fig. 4
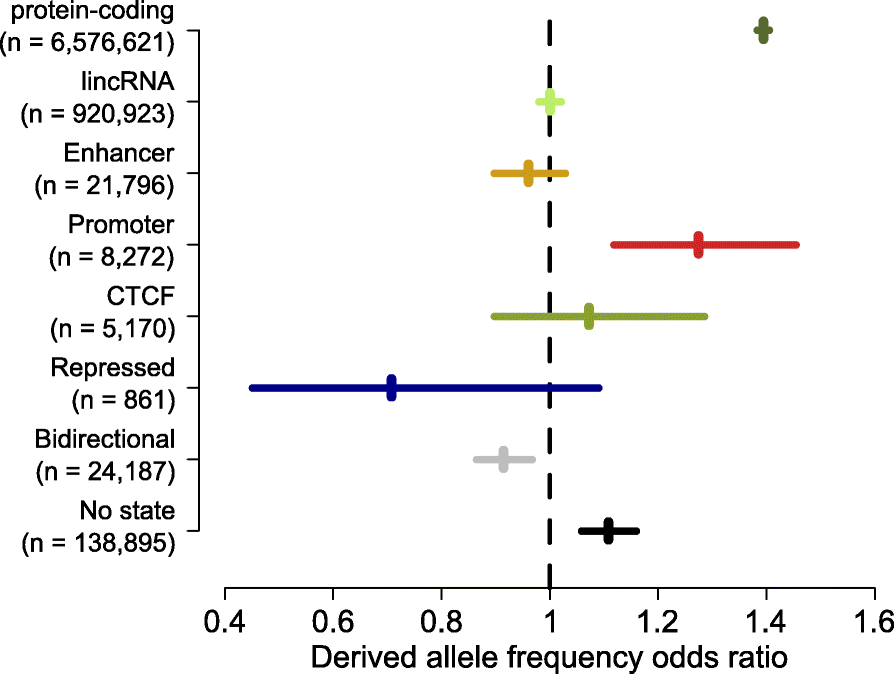
Mature eRNAs do not show signatures of selection within the human population. Odds ratios of frequencies within the deCODE population [49] for rare (<1.5%) and common (>5%) derived alleles compared between exonic and intronic sequences for transcripts overlapping the indicated genome annotations. The numbers of informative SNPs overlapping each category are shown in the parentheses next to the axis labels. Horizontal lines indicate the 95% confidence interval of the odds ratio estimates. Odds ratios significantly greater than one indicate increased selective constraint in exonic relative to intronic sequence
Similar articles
- Enhancer RNA: biogenesis, function, and regulation.
Ye R, Cao C, Xue Y. Ye R, et al. Essays Biochem. 2020 Dec 7;64(6):883-894. doi: 10.1042/EBC20200014. Essays Biochem. 2020. PMID: 33034351 Review. - Early adaptive chromatin remodeling events precede pathologic phenotypes and are reinforced in the failing heart.
Chapski DJ, Cabaj M, Morselli M, Mason RJ, Soehalim E, Ren S, Pellegrini M, Wang Y, Vondriska TM, Rosa-Garrido M. Chapski DJ, et al. J Mol Cell Cardiol. 2021 Nov;160:73-86. doi: 10.1016/j.yjmcc.2021.07.002. Epub 2021 Jul 15. J Mol Cell Cardiol. 2021. PMID: 34273410 Free PMC article. - Discovery of active enhancers through bidirectional expression of short transcripts.
Melgar MF, Collins FS, Sethupathy P. Melgar MF, et al. Genome Biol. 2011 Nov 14;12(11):R113. doi: 10.1186/gb-2011-12-11-r113. Genome Biol. 2011. PMID: 22082242 Free PMC article. - Enhancer identification in mouse embryonic stem cells using integrative modeling of chromatin and genomic features.
Chen CY, Morris Q, Mitchell JA. Chen CY, et al. BMC Genomics. 2012 Apr 26;13:152. doi: 10.1186/1471-2164-13-152. BMC Genomics. 2012. PMID: 22537144 Free PMC article. - Evaluating Enhancer Function and Transcription.
Field A, Adelman K. Field A, et al. Annu Rev Biochem. 2020 Jun 20;89:213-234. doi: 10.1146/annurev-biochem-011420-095916. Epub 2020 Mar 20. Annu Rev Biochem. 2020. PMID: 32197056 Review.
Cited by
- Enhancer activation from transposable elements in extrachromosomal DNA.
Kraft K, Murphy SE, Jones MG, Shi Q, Bhargava-Shah A, Luong C, Hung KL, He BJ, Li R, Park SK, Weiser NE, Luebeck J, Bafna V, Boeke JD, Mischel PS, Boettiger AN, Chang HY. Kraft K, et al. bioRxiv [Preprint]. 2024 Sep 8:2024.09.04.611262. doi: 10.1101/2024.09.04.611262. bioRxiv. 2024. PMID: 39282372 Free PMC article. Preprint. - The contribution of evolutionarily volatile promoters to molecular phenotypes and human trait variation.
Young RS, Talmane L, Marion de Procé S, Taylor MS. Young RS, et al. Genome Biol. 2022 Apr 4;23(1):89. doi: 10.1186/s13059-022-02634-w. Genome Biol. 2022. PMID: 35379293 Free PMC article. - Genome-wide RNA pol II initiation and pausing in neural progenitors of the rat.
Scheidegger A, Dunn CJ, Samarakkody A, Koney NK, Perley D, Saha RN, Nechaev S. Scheidegger A, et al. BMC Genomics. 2019 Jun 11;20(1):477. doi: 10.1186/s12864-019-5829-4. BMC Genomics. 2019. PMID: 31185909 Free PMC article. - Eukaryotic core promoters and the functional basis of transcription initiation.
Haberle V, Stark A. Haberle V, et al. Nat Rev Mol Cell Biol. 2018 Oct;19(10):621-637. doi: 10.1038/s41580-018-0028-8. Nat Rev Mol Cell Biol. 2018. PMID: 29946135 Free PMC article. Review. - Pervasive and CpG-dependent promoter-like characteristics of transcribed enhancers.
Steinhaus R, Gonzalez T, Seelow D, Robinson PN. Steinhaus R, et al. Nucleic Acids Res. 2020 Jun 4;48(10):5306-5317. doi: 10.1093/nar/gkaa223. Nucleic Acids Res. 2020. PMID: 32338759 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- MC_PC_U127527202/MRC_/Medical Research Council/United Kingdom
- MC_PC_U127527203/MRC_/Medical Research Council/United Kingdom
- MC_PC_U127597124/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous