Aerobic proteobacterial methylotrophs in Movile Cave: genomic and metagenomic analyses - PubMed (original) (raw)
Aerobic proteobacterial methylotrophs in Movile Cave: genomic and metagenomic analyses
Deepak Kumaresan et al. Microbiome. 2018.
Abstract
Background: Movile Cave (Mangalia, Romania) is a unique ecosystem where the food web is sustained by microbial primary production, analogous to deep-sea hydrothermal vents. Specifically, chemoautotrophic microbes deriving energy from the oxidation of hydrogen sulphide and methane form the basis of the food web.
Results: Here, we report the isolation of the first methane-oxidizing bacterium from the Movile Cave ecosystem, Candidatus Methylomonas sp. LWB, a new species and representative of Movile Cave microbial mat samples. While previous research has suggested a prevalence of anoxic conditions in deeper lake water and sediment, using small-scale shotgun metagenome sequencing, we show that metabolic genes encoding enzymes for aerobic methylotrophy are prevalent in sediment metagenomes possibly indicating the presence of microoxic conditions. Moreover, this study also indicates that members within the family Gallionellaceae (Sideroxydans and Gallionella) were the dominant taxa within the sediment microbial community, thus suggesting a major role for microaerophilic iron-oxidising bacteria in nutrient cycling within the Movile Cave sediments.
Conclusions: In this study, based on phylogenetic and metabolic gene surveys of metagenome sequences, the possibility of aerobic microbial processes (i.e., methylotrophy and iron oxidation) within the sediment is indicated. We also highlight significant gaps in our knowledge on biogeochemical cycles within the Movile Cave ecosystem, and the need to further investigate potential feedback mechanisms between microbial communities in both lake sediment and lake water.
Keywords: Extreme ecosystem; Methane; Methanotrophs; Methylotrophic bacteria; Movile cave; One-carbon metabolism.
Conflict of interest statement
Ethics approval
Not applicable
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Fig. 1
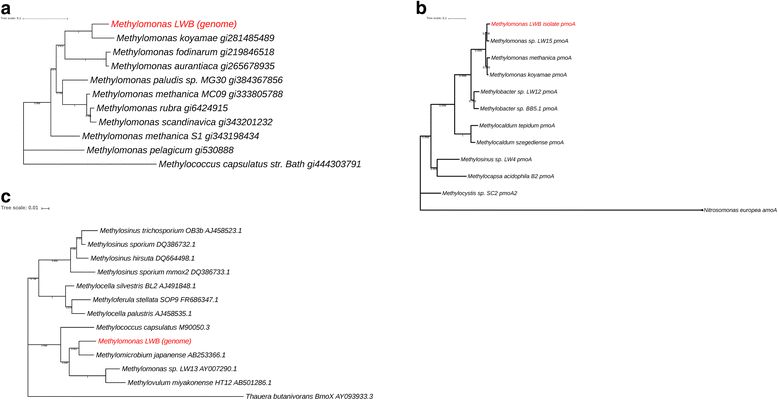
Phylogenetic neighbour-joining trees of (a) 16S rRNA gene sequences showing the relationship between the MOB isolate from Movile Cave to other members within the genus Methylomonas and Methylococcus capsulatus Bath. b Partial PmoA sequences amplified from the isolate LWB and other closely related PmoA sequences with the ammonia monooxygenase (AmoA) sequence as an out-group. Scale bar = 0.1 change per base position, (c) partial MmoX sequences derived from the MOB isolate genome and other closely related MmoX sequences with the butane monooxygenase (BmoX) sequence as an out-group. Scale bar = 0.01 change per base position
Fig. 2
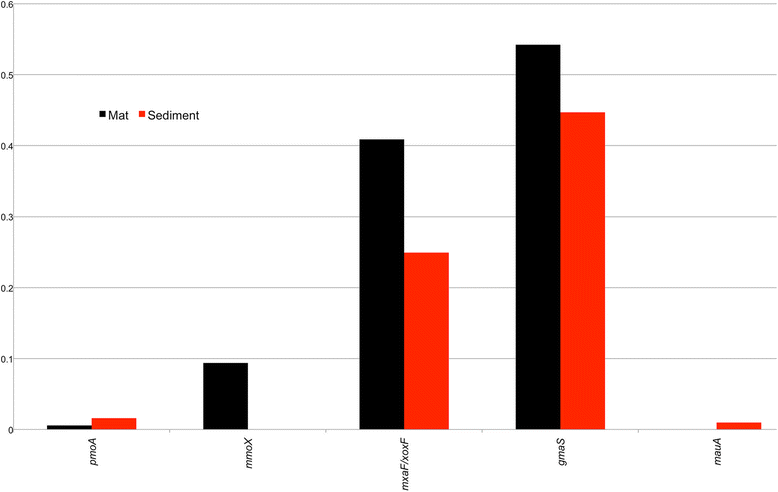
Comparison of relative abundance (normalized to RecA abundance) of different metabolic genes sequences in mat and sediment metagenomes. For one-carbon cycling: pmoA (particulate methane monooxygenase), mmox (soluble methane monoxygenase), mxaF/xoxF (methanol dehydrogenases), gmaS (gamma-glutamylmethylamide synthetase), mauA (methylamine dehydrogenase) and mcrA (methyl coenzyme M reductase)
Fig. 3
An approximately maximum-likelihood tree constructed using partial MmoX peptide sequences retrieved from the microbial mat metagenome, ratified MmoX peptide sequences (represented by blue font) and MmoX peptide sequence from the Methylomonas isolate from Movile Cave. Nodes with only MmoX peptide sequences from the microbial mat metagenomes are collapsed (grey triangles). Scale bar = 1 change per base position
Fig. 4
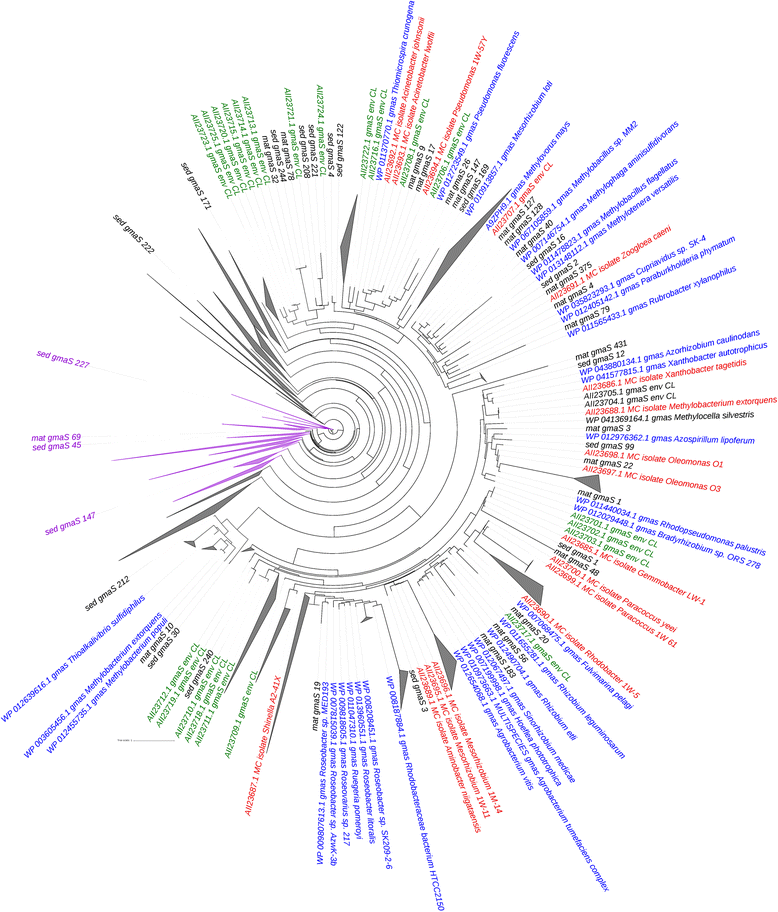
An approximately maximum-likelihood tree constructed using partial GmaS peptide sequences retrieved from both microbial mat and sediment metagenomes, ratified GmaS peptide sequences (represented in blue font), and both Movile Cave isolates (represented in red font) and environmental GmaS (represented in green font) peptide sequences. Nodes with GlnA (glutamine synthetase) peptide sequences are represented by purple colour. Nodes with only MxaF/XoxF peptide sequences from the microbial mat metagenomes are collapsed (triangles). Scale bar = 1 change per base position
Similar articles
- Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing.
Hutchens E, Radajewski S, Dumont MG, McDonald IR, Murrell JC. Hutchens E, et al. Environ Microbiol. 2004 Feb;6(2):111-20. doi: 10.1046/j.1462-2920.2003.00543.x. Environ Microbiol. 2004. PMID: 14756876 - Competition-cooperation in the chemoautotrophic ecosystem of Movile Cave: first metagenomic approach on sediments.
Chiciudean I, Russo G, Bogdan DF, Levei EA, Faur L, Hillebrand-Voiculescu A, Moldovan OT, Banciu HL. Chiciudean I, et al. Environ Microbiome. 2022 Aug 17;17(1):44. doi: 10.1186/s40793-022-00438-w. Environ Microbiome. 2022. PMID: 35978381 Free PMC article. - Life without light: microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave.
Chen Y, Wu L, Boden R, Hillebrand A, Kumaresan D, Moussard H, Baciu M, Lu Y, Colin Murrell J. Chen Y, et al. ISME J. 2009 Sep;3(9):1093-104. doi: 10.1038/ismej.2009.57. Epub 2009 May 28. ISME J. 2009. PMID: 19474813 - Methylotrophs in natural habitats: current insights through metagenomics.
Chistoserdova L. Chistoserdova L. Appl Microbiol Biotechnol. 2015 Jul;99(14):5763-79. doi: 10.1007/s00253-015-6713-z. Epub 2015 Jun 9. Appl Microbiol Biotechnol. 2015. PMID: 26051673 Review. - Modularity of methylotrophy, revisited.
Chistoserdova L. Chistoserdova L. Environ Microbiol. 2011 Oct;13(10):2603-22. doi: 10.1111/j.1462-2920.2011.02464.x. Epub 2011 Mar 28. Environ Microbiol. 2011. PMID: 21443740 Review.
Cited by
- A high-fibre personalised dietary advice given via a web tool reduces constipation complaints in adults.
Rijnaarts I, de Roos NM, Wang T, Zoetendal EG, Top J, Timmer M, Hogenelst K, Bouwman EP, Witteman B, de Wit N. Rijnaarts I, et al. J Nutr Sci. 2022 Apr 28;11:e31. doi: 10.1017/jns.2022.27. eCollection 2022. J Nutr Sci. 2022. PMID: 35573462 Free PMC article. - Microorganisms as bio-filters to mitigate greenhouse gas emissions from high-altitude permafrost revealed by nanopore-based metagenomics.
Dang C, Wu Z, Zhang M, Li X, Sun Y, Wu R, Zheng Y, Xia Y. Dang C, et al. Imeta. 2022 May 8;1(2):e24. doi: 10.1002/imt2.24. eCollection 2022 Jun. Imeta. 2022. PMID: 38868568 Free PMC article. - Metal(loid) speciation and transformation by aerobic methanotrophs.
Karthikeyan OP, Smith TJ, Dandare SU, Parwin KS, Singh H, Loh HX, Cunningham MR, Williams PN, Nichol T, Subramanian A, Ramasamy K, Kumaresan D. Karthikeyan OP, et al. Microbiome. 2021 Jul 6;9(1):156. doi: 10.1186/s40168-021-01112-y. Microbiome. 2021. PMID: 34229757 Free PMC article. Review. - Microbial Interactions Drive Distinct Taxonomic and Potential Metabolic Responses to Habitats in Karst Cave Ecosystem.
Ma L, Huang X, Wang H, Yun Y, Cheng X, Liu D, Lu X, Qiu X. Ma L, et al. Microbiol Spectr. 2021 Oct 31;9(2):e0115221. doi: 10.1128/Spectrum.01152-21. Epub 2021 Sep 8. Microbiol Spectr. 2021. PMID: 34494852 Free PMC article. - metaGOflow: a workflow for the analysis of marine Genomic Observatories shotgun metagenomics data.
Zafeiropoulos H, Beracochea M, Ninidakis S, Exter K, Potirakis A, De Moro G, Richardson L, Corre E, Machado J, Pafilis E, Kotoulas G, Santi I, Finn RD, Cox CJ, Pavloudi C. Zafeiropoulos H, et al. Gigascience. 2022 Dec 28;12:giad078. doi: 10.1093/gigascience/giad078. Epub 2023 Oct 18. Gigascience. 2022. PMID: 37850871 Free PMC article.
References
- Sarbu SM, Vlasceanu L, Popa R, Sheridan P, Kinkle BK, Kane TC. Microbial mats in a thermomineral sulfurous cave. Berlin: Springer-Verlag; 1994.
- Kumaresan D, Wischer D, Stephenson J, Hillebrand-Voiculescu A, Murrell JC. Microbiology of Movile cave—a chemolithoautotrophic ecosystem. Geomicrobiol J. 2014;31:186–193. doi: 10.1080/01490451.2013.839764. - DOI
- Sarbu SM. Movile cave: a chemoautotrophically based ground water ecosystem. In: Wilkens H, Culver DC, Humphreys WF, editors. Subterranean ecosystems. Amsterdan: Elsevier; 2000. pp. 319–343.
- Muschiol D, Giere O, Transpurger W. Population dynamics of a cavernicolous nematode community in a chemoautotrophic groundwater system. Limnol Oceanogr. 2015;60:127–135. doi: 10.1002/lno.10017. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources