Extreme haplotype variation in the desiccation-tolerant clubmoss Selaginella lepidophylla - PubMed (original) (raw)
Extreme haplotype variation in the desiccation-tolerant clubmoss Selaginella lepidophylla
Robert VanBuren et al. Nat Commun. 2018.
Abstract
Plant genome size varies by four orders of magnitude, and most of this variation stems from dynamic changes in repetitive DNA content. Here we report the small 109 Mb genome of Selaginella lepidophylla, a clubmoss with extreme desiccation tolerance. Single-molecule sequencing enables accurate haplotype assembly of a single heterozygous S. lepidophylla plant, revealing extensive structural variation. We observe numerous haplotype-specific deletions consisting of largely repetitive and heavily methylated sequences, with enrichment in young Gypsy LTR retrotransposons. Such elements are active but rapidly deleted, suggesting "bloat and purge" to maintain a small genome size. Unlike all other land plant lineages, Selaginella has no evidence of a whole-genome duplication event in its evolutionary history, but instead shows unique tandem gene duplication patterns reflecting adaptation to extreme drying. Gene expression changes during desiccation in S. lepidophylla mirror patterns observed across angiosperm resurrection plants.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
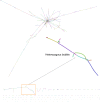
Fig. 1
Genome assembly graph of S. lepidophylla. Each line (node) represents a contig with connections (edges) representing ambiguities in the graph structure. A subset of the graph showing heterozygous bubbles is enlarged. Contig color is randomly assigned
Fig. 2
Extensive haplotype-specific LTR retrotransposon accumulation and deletion. a Typical micro-collinearity between two S. lepidophylla haplotypes. b Estimated insertions times of intact Copia and Gypsy LTR retrotransposons. c Composition of haplotype specific and paired collinear regions. The proportion of genes and LTRs in the 105 manually curated insertions (left) and flanking collinear regions (right) are plotted. d Micro-collinearity between two haplotypes showing a large 57 kb deletion in contig 142. Green and blue bars delineate gene orientation. Predicted LTR elements are depicted in orange and the three complete Gypsy LTRs are denoted by arrows. The three complete Gypsy LTRs have insertion times of <0.1 Ma
Fig. 3
Global methylation patterns in the S. lepidophylla genome. a S. lepidophylla lacks gene body methylation. Levels of CHH (green), CHG (blue) and CpG (red) methylation are plotted in the upstream (transcriptional start site; TSS), downstream (transcriptional termination site (TTS), and body of genes. b Intact LTR retrotransposons are highly methylated with decreasing levels at the 5′- and 3′-flanking regions. c Methylation levels are highly variable across the genome with strong correlation of LTR retrotransposon density. Top, genome landscape of genomic features across Contig 1. Bottom, heatmap of genomic features and methylation levels, where blue indicates low abundance and red signifies high abundance
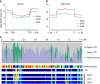
Fig. 4
Genomic features of the rehydration and desiccation processes. a Overview of sampling for the rehydration and desiccation time course. Samples were taken from plants that were desiccated for 3 years (0), 1, 6, and 24 h post rehydration (recovery), 120 h post rehydration (fully recovered), and after a 24 h dehydration (de24 h). b Scaled transcript expression profiles (in transcripts per million, TPM) of representative gene co-expression clusters with decreased (10, 16, 34) and increased expression (5, 14, 34) during rehydration. c Heatmap of log2 transformed early light-induced protein (ELIP) expression with the two arrays of tandem duplicated genes in S. lepidophylla orthologous to the only ELIP in to S. moellendorffii highlighted on the left
Similar articles
- The desiccation tolerant secrets of Selaginella lepidophylla: what we have learned so far?
Pampurova S, Van Dijck P. Pampurova S, et al. Plant Physiol Biochem. 2014 Jul;80:285-90. doi: 10.1016/j.plaphy.2014.04.015. Epub 2014 Apr 26. Plant Physiol Biochem. 2014. PMID: 24813728 - Comparative metabolic profiling between desiccation-sensitive and desiccation-tolerant species of Selaginella reveals insights into the resurrection trait.
Yobi A, Wone BW, Xu W, Alexander DC, Guo L, Ryals JA, Oliver MJ, Cushman JC. Yobi A, et al. Plant J. 2012 Dec;72(6):983-99. doi: 10.1111/tpj.12008. Epub 2012 Oct 19. Plant J. 2012. PMID: 23061970 - Genome Analysis of the Ancient Tracheophyte Selaginella tamariscina Reveals Evolutionary Features Relevant to the Acquisition of Desiccation Tolerance.
Xu Z, Xin T, Bartels D, Li Y, Gu W, Yao H, Liu S, Yu H, Pu X, Zhou J, Xu J, Xi C, Lei H, Song J, Chen S. Xu Z, et al. Mol Plant. 2018 Jul 2;11(7):983-994. doi: 10.1016/j.molp.2018.05.003. Epub 2018 May 17. Mol Plant. 2018. PMID: 29777775 - Molecular mechanisms of desiccation tolerance in resurrection plants.
Gechev TS, Dinakar C, Benina M, Toneva V, Bartels D. Gechev TS, et al. Cell Mol Life Sci. 2012 Oct;69(19):3175-86. doi: 10.1007/s00018-012-1088-0. Epub 2012 Jul 26. Cell Mol Life Sci. 2012. PMID: 22833170 Free PMC article. Review. - Programming desiccation-tolerance: from plants to seeds to resurrection plants.
Farrant JM, Moore JP. Farrant JM, et al. Curr Opin Plant Biol. 2011 Jun;14(3):340-5. doi: 10.1016/j.pbi.2011.03.018. Epub 2011 Apr 19. Curr Opin Plant Biol. 2011. PMID: 21511516 Review.
Cited by
- Nanoparticle-Mediated Genetic Transformation in a Selaginella Species.
Ariyarathne MA, Wone B, Wijewantha N, Wone BWM. Ariyarathne MA, et al. Genes (Basel). 2024 Aug 19;15(8):1091. doi: 10.3390/genes15081091. Genes (Basel). 2024. PMID: 39202450 Free PMC article. - The Possible Earliest Allopolyploidization in Tracheophytes Revealed by Phylotranscriptomics and Morphology of Selaginellaceae.
Kang JS, Yu JG, Xiang QP, Zhang XC. Kang JS, et al. Mol Biol Evol. 2024 Aug 2;41(8):msae153. doi: 10.1093/molbev/msae153. Mol Biol Evol. 2024. PMID: 39101470 Free PMC article. - Convergent evolution of desiccation tolerance in grasses.
Marks RA, Van Der Pas L, Schuster J, Gilman IS, VanBuren R. Marks RA, et al. Nat Plants. 2024 Jul;10(7):1112-1125. doi: 10.1038/s41477-024-01729-5. Epub 2024 Jun 21. Nat Plants. 2024. PMID: 38906996 - Extraordinary preservation of gene collinearity over three hundred million years revealed in homosporous lycophytes.
Li C, Wickell D, Kuo LY, Chen X, Nie B, Liao X, Peng D, Ji J, Jenkins J, Williams M, Shu S, Plott C, Barry K, Rajasekar S, Grimwood J, Han X, Sun S, Hou Z, He W, Dai G, Sun C, Schmutz J, Leebens-Mack JH, Li FW, Wang L. Li C, et al. Proc Natl Acad Sci U S A. 2024 Jan 23;121(4):e2312607121. doi: 10.1073/pnas.2312607121. Epub 2024 Jan 18. Proc Natl Acad Sci U S A. 2024. PMID: 38236735 Free PMC article. - Utilizing transcriptomics and metabolomics to unravel key genes and metabolites of maize seedlings in response to drought stress.
Li Y, Su Z, Lin Y, Xu Z, Bao H, Wang F, Liu J, Hu S, Wang Z, Yu X, Gao J. Li Y, et al. BMC Plant Biol. 2024 Jan 8;24(1):34. doi: 10.1186/s12870-023-04712-y. BMC Plant Biol. 2024. PMID: 38185653 Free PMC article.
References
- Oliver MJ, Tuba Z, Mishler BD. The evolution of vegetative desiccation tolerance in land plants. Plant Ecol. 2000;151:85–100. doi: 10.1023/A:1026550808557. - DOI
- Proctor M. The physiological basis of bryophyte production. Bot. J. Linn. Soc. 1990;104:61–77. doi: 10.1111/j.1095-8339.1990.tb02211.x. - DOI
- Lüttge, U., Beck, E. & Bartels, D. Plant desiccation tolerance. Vol. 215 (Springer, 2011).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous