Analysis of the codon usage pattern in Middle East Respiratory Syndrome Coronavirus - PubMed (original) (raw)
Analysis of the codon usage pattern in Middle East Respiratory Syndrome Coronavirus
Ye Chen et al. Oncotarget. 2017.
Abstract
Middle East Respiratory Syndrome Coronavirus (MERS-CoV), which first broken out in Jeddah in 2012, causes a severe acute respiratory illness with a high mortality rate. To better understand the molecular characteristics of isolated MERS-CoV genomes, we first analysed the codon usage pattern of the zoonotic MERS-CoV strains comprehensively to gain an insight into the mechanism of cross-species transmission. We found that MERS human/camel isolates showed a low codon usage bias. Both mutation and nature selection pressure have contributed to this low codon usage bias, with the former being the main determining factor. We also observed that gene function, evolution time and the different host species of the virus all contributed to the bias of MERS-CoV, to some extent. Additionally, the codon usage pattern of MERS-CoV isolates is different from other related Nidovirales viruses isolated from bats and hedgehogs. In the future, more epidemiological surveys are required to examine the factors that resulted in the emergence and outbreak of this virus.
Keywords: MERS-CoV; codon usage pattern; mutation bias; natural selection.
Conflict of interest statement
CONFLICTS OF INTEREST Competing financial interests: The authors declare no competing financial interests.
Figures
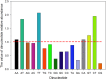
Figure 1. The relative abundance values of the 16 dinucleotides
The different colours represent the different dinucleotides. The red dotted line indicates that the relative abundance value of a dinucleotide is 1.
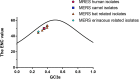
Figure 2. The plots of ENC values against GC3s values for MERS-CoV and MERS-CoV related strains
All the points corresponding to human, camel isolated MERS-CoV strains and bat and hedgehog(erinaceus) isolated CoV were labelled in circle, square, triangle, and rhombus, respectively.
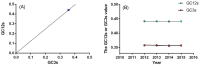
Figure 3
(A) The neutral analysis of GC3s against GC12s. (B) The evolutionary analysis of the GC3s and GC12s values. The solid line represents the regression line.
Figure 4. CA of MERS-CoV human/camel isolates
Each viral gene is displayed in a 3-dimensional representation. The X, Y and Z-axes have arbitrary scales generated by the CA and the weight of each codon in these axes varies in different segments. The codon usage trends with time of the viral isolates are indicated by different colours. The different hosts of the MRES-CoV isolates are indicated by different shapes. (A), (B), (C), (D) and (E) represents the 3D graph of the M, N, ORF1ab, S and the complete genome, respectively, using the CA data.
Similar articles
- Comparative Genomic Analysis MERS CoV Isolated from Humans and Camels with Special Reference to Virus Encoded Helicase.
Alnazawi M, Altaher A, Kandeel M. Alnazawi M, et al. Biol Pharm Bull. 2017;40(8):1289-1298. doi: 10.1248/bpb.b17-00241. Biol Pharm Bull. 2017. PMID: 28769010 - Analysis of Codon Usage and Nucleotide Bias in Middle East Respiratory Syndrome Coronavirus Genes.
Hussain S, Shinu P, Islam MM, Chohan MS, Rasool ST. Hussain S, et al. Evol Bioinform Online. 2020 May 4;16:1176934320918861. doi: 10.1177/1176934320918861. eCollection 2020. Evol Bioinform Online. 2020. PMID: 32425493 Free PMC article. - Synonymous and Biased Codon Usage by MERS CoV Papain-Like and 3CL-Proteases.
Kandeel M, Altaher A. Kandeel M, et al. Biol Pharm Bull. 2017 Jul 1;40(7):1086-1091. doi: 10.1248/bpb.b17-00168. Epub 2017 Apr 18. Biol Pharm Bull. 2017. PMID: 28420819 - Genomics and zoonotic infections: Middle East respiratory syndrome.
Wernery U, Lau SK, Woo PC. Wernery U, et al. Rev Sci Tech. 2016 Apr;35(1):191-202. doi: 10.20506/rst.35.1.2427. Rev Sci Tech. 2016. PMID: 27217178 Review. - Coronavirus Host Range Expansion and Middle East Respiratory Syndrome Coronavirus Emergence: Biochemical Mechanisms and Evolutionary Perspectives.
Peck KM, Burch CL, Heise MT, Baric RS. Peck KM, et al. Annu Rev Virol. 2015 Nov;2(1):95-117. doi: 10.1146/annurev-virology-100114-055029. Epub 2015 Aug 7. Annu Rev Virol. 2015. PMID: 26958908 Review.
Cited by
- Understanding the origin of COVID-19 requires to change the paradigm on zoonotic emergence from the spillover to the circulation model.
Frutos R, Gavotte L, Devaux CA. Frutos R, et al. Infect Genet Evol. 2021 Nov;95:104812. doi: 10.1016/j.meegid.2021.104812. Epub 2021 Mar 18. Infect Genet Evol. 2021. PMID: 33744401 Free PMC article. Review. - Codon Usage and Phenotypic Divergences of SARS-CoV-2 Genes.
Dilucca M, Forcelloni S, Georgakilas AG, Giansanti A, Pavlopoulou A. Dilucca M, et al. Viruses. 2020 Apr 30;12(5):498. doi: 10.3390/v12050498. Viruses. 2020. PMID: 32366025 Free PMC article. - Deciphering the co-adaptation of codon usage between respiratory coronaviruses and their human host uncovers candidate therapeutics for COVID-19.
Nambou K, Anakpa M. Nambou K, et al. Infect Genet Evol. 2020 Nov;85:104471. doi: 10.1016/j.meegid.2020.104471. Epub 2020 Jul 22. Infect Genet Evol. 2020. PMID: 32707288 Free PMC article. - CoronaVR: A Computational Resource and Analysis of Epitopes and Therapeutics for Severe Acute Respiratory Syndrome Coronavirus-2.
Gupta AK, Khan MS, Choudhury S, Mukhopadhyay A, Sakshi, Rastogi A, Thakur A, Kumari P, Kaur M, Shalu, Saini C, Sapehia V, Barkha, Patel PK, Bhamare KT, Kumar M. Gupta AK, et al. Front Microbiol. 2020 Jul 31;11:1858. doi: 10.3389/fmicb.2020.01858. eCollection 2020. Front Microbiol. 2020. PMID: 32849449 Free PMC article. - Capturing a Crucial 'Disorder-to-Order Transition' at the Heart of the Coronavirus Molecular Pathology-Triggered by Highly Persistent, Interchangeable Salt-Bridges.
Roy S, Ghosh P, Bandyopadhyay A, Basu S. Roy S, et al. Vaccines (Basel). 2022 Feb 16;10(2):301. doi: 10.3390/vaccines10020301. Vaccines (Basel). 2022. PMID: 35214759 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources