Hippo pathway coactivators Yap and Taz are required to coordinate mammalian liver regeneration - PubMed (original) (raw)
Hippo pathway coactivators Yap and Taz are required to coordinate mammalian liver regeneration
Li Lu et al. Exp Mol Med. 2018.
Abstract
The mammalian liver has a remarkable capacity for repair following injury. Removal of up to two-third of liver mass results in a series of events that include extracellular matrix remodeling, coordinated hepatic cell cycle re-entry, restoration of liver mass and tissue remodeling to return the damaged liver to its normal state. Although there has been considerable advancement of our knowledge concerning the regenerative capacity of the mammalian liver, many outstanding questions remaining, such as: how does the regenerating liver stop proliferating when appropriate mass is restored and how do these mechanisms relate to normal regulation of organ size during development? Hippo pathway has been proposed to be central in mediating both events: organ size control during development and following regeneration. In this report, we examined the role of Yap and Taz, key components of the Hippo pathway in liver organ size regulation, both in the context of development and homeostasis. Our studies reveal that contrary to the current paradigms that Yap/Taz are not required for developmental regulation of liver size but are required for proper liver regeneration. In livers depleted of Yap and Taz, liver mass is elevated in neonates and adults. However, Yap/Taz-depleted livers exhibit profound defects in liver regeneration, including an inability to restore liver mass and to properly coordinate cell cycle entry. Taken together, our results highlight requirements for the Hippo pathway during liver regeneration and indicate that there are additional pathways that cooperate with Hippo signaling to control liver size during development and in the adult.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Figure 1
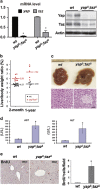
Efficient deletion of Yap and Taz in the mouse liver. (a) Both mRNA and protein levels of the Yap and Taz are significantly reduced in _yap_Δ/_taz_Δ liver. (b) _Yap_Δ/_taz_Δ liver is ~20% larger than wild-type (wt) counterparts at 2 month old and ~50% larger at 1 year old. (c) Hepatic macrophages in area of necrosis in 2-month-old _yap_Δ/_taz_Δ liver suggests hepatic injury. (d) Elevated serum AST and ALT levels in 2-month-old _yap_Δ/_taz_Δ liver indicate hepatic injury by yap/taz deletion. _N_=3. (e) Compensatory hepatic proliferation evidenced by significantly increased BrdU incorporation in 2-month-old _yap_Δ/_taz_Δ liver. _N_=3. *P<0.05.
Figure 2
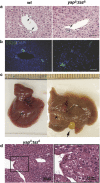
Inflammation, biliary tract defects and adenoma formation in _yap_Δ/_taz_Δ livers. (a) Inflammation around bile ducts in _yap_Δ/_taz_Δ livers seen by H&E histology. Scale bar is 50 μm. (b) The irregularly shaped and less well-formed bile ducts in _yap_Δ/_taz_Δ livers distinguished by CK19 staining. Scale bar is 50 μm. (c) Gross view of a 1 year old wild-type liver and a _yap_Δ/_taz_Δ liver with adenoma (star labeled) and enlarged gallbladder (arrows pointed). (d) The liver adenoma histology of low-power (left) and high-power (right) in 1 year old _yap_Δ/_taz_Δ liver.
Figure 3
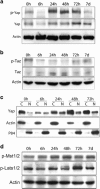
Regulation of Hippo signaling during liver regeneration. (a) Yap phosphorylation transiently decreases after partial hepatectomy (PHx), suggesting Yap activation during liver regeneration. (b) Taz phosphorylation also decreases after PHx, suggesting Taz activation during liver regeneration. (c) Nuclear and cytoplasmic localization of Yap after PHx by western analysis. (d) Phosphorylation activations of Mst1/2 and Lats1/2 after PHx are not seen. Results were confirmed in three independently repeated experiments.
Figure 4
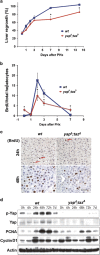
Inefficient liver regeneration in _yap_Δ/_taz_Δ mice. (a) Liver regrowth is blunted in _yap_Δ/_taz_Δ liver at all time points. (b) Diminished entry of _yap_Δ/_taz_Δ hepatocytes in to S-phase as assayed by BrdU incorporation. (c) Represented images of BrdU staining at 24 and 48 h after partial hepatectomy. Scale bar is 50 μm. (d) Deficient hepatic cell cycle re-entry and progression in _yap_Δ/_taz_Δ liver by western analysis of the cell cycle markers. *P<0.05. Results were confirmed in three independently repeated experiments.
Comment in
- 5-HT and Intraplatelet 5-HT: a potential upstream regulator of YAP in liver regeneration.
He C, Zhai M, Shu B, Deng C, Li L, Liu S. He C, et al. Exp Mol Med. 2019 Oct 16;51(10):1-2. doi: 10.1038/s12276-019-0324-1. Exp Mol Med. 2019. PMID: 31619665 Free PMC article. No abstract available.
Similar articles
- Proliferation of hepatic stellate cells, mediated by YAP and TAZ, contributes to liver repair and regeneration after liver ischemia-reperfusion injury.
Konishi T, Schuster RM, Lentsch AB. Konishi T, et al. Am J Physiol Gastrointest Liver Physiol. 2018 Apr 1;314(4):G471-G482. doi: 10.1152/ajpgi.00153.2017. Epub 2018 Jan 11. Am J Physiol Gastrointest Liver Physiol. 2018. PMID: 29351389 Free PMC article. - WWC1 and NF2 Prevent the Development of Intrahepatic Cholangiocarcinoma by Regulating YAP/TAZ Activity through LATS in Mice.
Park J, Kim JS, Nahm JH, Kim SK, Lee DH, Lim DS. Park J, et al. Mol Cells. 2020 May 31;43(5):491-499. doi: 10.14348/molcells.2020.0093. Mol Cells. 2020. PMID: 32451369 Free PMC article. - A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis.
Moroishi T, Park HW, Qin B, Chen Q, Meng Z, Plouffe SW, Taniguchi K, Yu FX, Karin M, Pan D, Guan KL. Moroishi T, et al. Genes Dev. 2015 Jun 15;29(12):1271-84. doi: 10.1101/gad.262816.115. Genes Dev. 2015. PMID: 26109050 Free PMC article. - Physiological and pathological roles of the Hippo-YAP/TAZ signaling pathway in liver formation, homeostasis, and tumorigenesis.
Nishina H. Nishina H. Cancer Sci. 2022 Jun;113(6):1900-1908. doi: 10.1111/cas.15352. Epub 2022 Apr 17. Cancer Sci. 2022. PMID: 35349740 Free PMC article. Review. - Insights into the Regulation of Yap/Taz from Cellular Systems and Mouse Models.
Du W, Du W, Wan M, Zhou X, Xu X, Zheng L. Du W, et al. Curr Stem Cell Res Ther. 2018;13(1):16-25. doi: 10.2174/1574888X12666170102124421. Curr Stem Cell Res Ther. 2018. PMID: 28042766 Review.
Cited by
- The essential role of TAZ in normal tissue homeostasis.
Jeong MG, Kim HK, Hwang ES. Jeong MG, et al. Arch Pharm Res. 2021 Mar;44(3):253-262. doi: 10.1007/s12272-021-01322-w. Epub 2021 Mar 26. Arch Pharm Res. 2021. PMID: 33770379 Free PMC article. Review. - Decoding YAP dependent transcription in the liver.
Biagioni F, Croci O, Sberna S, Donato E, Sabò A, Bisso A, Curti L, Chiesa A, Campaner S. Biagioni F, et al. Nucleic Acids Res. 2022 Aug 12;50(14):7959-7971. doi: 10.1093/nar/gkac624. Nucleic Acids Res. 2022. PMID: 35871292 Free PMC article. - YAP and TAZ Heterogeneity in Primary Liver Cancer: An Analysis of Its Prognostic and Diagnostic Role.
Van Haele M, Moya IM, Karaman R, Rens G, Snoeck J, Govaere O, Nevens F, Verslype C, Topal B, Monbaliu D, Halder G, Roskams T. Van Haele M, et al. Int J Mol Sci. 2019 Feb 1;20(3):638. doi: 10.3390/ijms20030638. Int J Mol Sci. 2019. PMID: 30717258 Free PMC article. - Bile canaliculi remodeling activates YAP via the actin cytoskeleton during liver regeneration.
Meyer K, Morales-Navarrete H, Seifert S, Wilsch-Braeuninger M, Dahmen U, Tanaka EM, Brusch L, Kalaidzidis Y, Zerial M. Meyer K, et al. Mol Syst Biol. 2020 Feb;16(2):e8985. doi: 10.15252/msb.20198985. Mol Syst Biol. 2020. PMID: 32090478 Free PMC article. - Hippo Signaling Controls NLR Family Pyrin Domain Containing 3 Activation and Governs Immunoregulation of Mesenchymal Stem Cells in Mouse Liver Injury.
Li C, Jin Y, Wei S, Sun Y, Jiang L, Zhu Q, Farmer DG, Busuttil RW, Kupiec-Weglinski JW, Ke B. Li C, et al. Hepatology. 2019 Nov;70(5):1714-1731. doi: 10.1002/hep.30700. Epub 2019 Jun 21. Hepatology. 2019. PMID: 31063235 Free PMC article.
References
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol 2007; 17: 2054–2060. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases