Machine learning for identifying Randomized Controlled Trials: An evaluation and practitioner's guide - PubMed (original) (raw)
Machine learning for identifying Randomized Controlled Trials: An evaluation and practitioner's guide
Iain J Marshall et al. Res Synth Methods. 2018 Dec.
Abstract
Machine learning (ML) algorithms have proven highly accurate for identifying Randomized Controlled Trials (RCTs) but are not used much in practice, in part because the best way to make use of the technology in a typical workflow is unclear. In this work, we evaluate ML models for RCT classification (support vector machines, convolutional neural networks, and ensemble approaches). We trained and optimized support vector machine and convolutional neural network models on the titles and abstracts of the Cochrane Crowd RCT set. We evaluated the models on an external dataset (Clinical Hedges), allowing direct comparison with traditional database search filters. We estimated area under receiver operating characteristics (AUROC) using the Clinical Hedges dataset. We demonstrate that ML approaches better discriminate between RCTs and non-RCTs than widely used traditional database search filters at all sensitivity levels; our best-performing model also achieved the best results to date for ML in this task (AUROC 0.987, 95% CI, 0.984-0.989). We provide practical guidance on the role of ML in (1) systematic reviews (high-sensitivity strategies) and (2) rapid reviews and clinical question answering (high-precision strategies) together with recommended probability cutoffs for each use case. Finally, we provide open-source software to enable these approaches to be used in practice.
© 2018 The Authors. Research Synthesis Methods published by John Wiley & Sons Ltd.
Figures
Figure 1
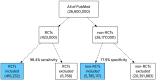
Tree diagram: The false positive burden associated with using a high‐sensitivity search compounded by RCTs being a minority class. Illustrative figures, assuming that 1.6% of all articles are RCTs (based on PubMed search; approximately 423 000 in total), and a search filter with 98.4% sensitivity and 77.9% specificity (the performance of the Cochrane HSSS based on data from McKibbon et al9). The 2 blue shaded boxes together represent the search retrieval. The search filter thus retrieves a total of 6 201 349 articles, of which only 416 232 (or 6.7%) are actually RCTs (being the precision statistic) [Colour figure can be viewed at
wileyonlinelibrary.com
]
Figure 2
Receiver operating characteristic scatterplot for conventional database filters (based on data published by McKibbon et al,9 with the 2 comparator strategies from this analysis labeled. RCT PT tag, the single‐term strategy based on the manually applied PT tag (the high‐precision comparator); Cochrane HSSS, the Cochrane Highly Sensitive Search Strategy (the high‐sensitivity comparator) [Colour figure can be viewed at
wileyonlinelibrary.com
]
Figure 3
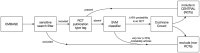
The Cochrane Crowd/EMBASE project pipeline. Source articles (titles and abstracts) are identified via a sensitive database search filter. Articles already tagged as being RCTs (via Emtree PT tag) are sent directly to CENTRAL. Articles predicted to have <10% probability of being RCTs via an SVM classifier are directly excluded. The remaining articles are considered by the crowd
Figure 4
Schematic illustrating separating plane in support vector machines, here depicted in 2 dimensions. Here, the separating plane (a straight line in this two‐dimensional case) is depicted as the black line and the margin are depicted in gray. The instances nearest to the margin (support vectors) are highlighted in white
Figure 5
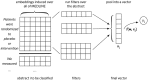
Schematic illustrating convolutional neural network architecture for text classification. Here, y i is the label (RCT or not) for document i, w is a weight vector associated with the classification layer, and x i is the vector representation of document i induced by the CNN
Figure 6
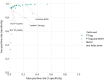
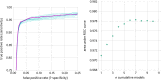
Receiver operating characteristics of the machine learning algorithms trained on plain text alone, (1) support vector machine, (2) convolutional neural network both single model, and bagged result of 10 models (each trained on all RCTs and a different random sample of non‐RCTs). The points depict the 3 conventional database filters, which use plain text only and do not require use of MeSH/PT tags. The blue shaded area in the left part of the figure is enlarged on the right‐side bottom section
Figure 7
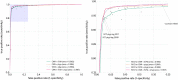
Left: Receiver operating characteristics curve (zoomed to accentuate variance); effects of balanced sampling: The individual models are depicted in light blue; the magenta curve depicts the performance of the consensus classification (the mean probability score of being an RCT from the component models). Right: Cumulative performance (area under receiver operating characteristics curve) of bagging multiple models trained on balanced samples. Performance increases until approximately 6 models are included, then is static afterwards
Figure 8
Receiver operating characteristics curve: Hybrid/ensembled models including use of the manually applied PT tag. The area bounded by the blue shaded area on the left‐hand plot is enlarged on the right to illustrate differences between models and conventional database filters. Note that the RCT PT tag has become more sensitive from 20099 to 2017 (the reanalysis conducted here), reflecting the late application of the tag to missed RCTs including through data provided to PubMed by the Cochrane Collaboration11
Figure 9
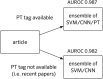
PubMed PT information is used where present; where not, the best‐performing text‐alone approach is automatically used, with a modest reduction in accuracy
Similar articles
- Machine learning reduced workload with minimal risk of missing studies: development and evaluation of a randomized controlled trial classifier for Cochrane Reviews.
Thomas J, McDonald S, Noel-Storr A, Shemilt I, Elliott J, Mavergames C, Marshall IJ. Thomas J, et al. J Clin Epidemiol. 2021 May;133:140-151. doi: 10.1016/j.jclinepi.2020.11.003. Epub 2020 Nov 7. J Clin Epidemiol. 2021. PMID: 33171275 Free PMC article. - Identifying reports of randomized controlled trials (RCTs) via a hybrid machine learning and crowdsourcing approach.
Wallace BC, Noel-Storr A, Marshall IJ, Cohen AM, Smalheiser NR, Thomas J. Wallace BC, et al. J Am Med Inform Assoc. 2017 Nov 1;24(6):1165-1168. doi: 10.1093/jamia/ocx053. J Am Med Inform Assoc. 2017. PMID: 28541493 Free PMC article. - Machine learning algorithms to identify cluster randomized trials from MEDLINE and EMBASE.
Al-Jaishi AA, Taljaard M, Al-Jaishi MD, Abdullah SS, Thabane L, Devereaux PJ, Dixon SN, Garg AX. Al-Jaishi AA, et al. Syst Rev. 2022 Oct 25;11(1):229. doi: 10.1186/s13643-022-02082-4. Syst Rev. 2022. PMID: 36284336 Free PMC article. - Identifying additional studies for a systematic review of retention strategies in randomised controlled trials: making contact with trials units and trial methodologists.
Brueton V, Tierney JF, Stenning S, Rait G. Brueton V, et al. Syst Rev. 2017 Aug 22;6(1):167. doi: 10.1186/s13643-017-0549-9. Syst Rev. 2017. PMID: 28830570 Free PMC article. Review. - Use of methodological search filters to identify diagnostic accuracy studies can lead to the omission of relevant studies.
Leeflang MM, Scholten RJ, Rutjes AW, Reitsma JB, Bossuyt PM. Leeflang MM, et al. J Clin Epidemiol. 2006 Mar;59(3):234-40. doi: 10.1016/j.jclinepi.2005.07.014. J Clin Epidemiol. 2006. PMID: 16488353 Review.
Cited by
- Semi-automated title-abstract screening using natural language processing and machine learning.
Pilz M, Zimmermann S, Friedrichs J, Wördehoff E, Ronellenfitsch U, Kieser M, Vey JA. Pilz M, et al. Syst Rev. 2024 Nov 1;13(1):274. doi: 10.1186/s13643-024-02688-w. Syst Rev. 2024. PMID: 39487499 Free PMC article. - Nasal continuous positive airway pressure immediately after extubation for preventing morbidity in preterm infants.
Ho JJ, Kidman AM, Chua B, Chang G, Fiander M, Davis PG. Ho JJ, et al. Cochrane Database Syst Rev. 2024 Oct 11;10(10):CD000143. doi: 10.1002/14651858.CD000143.pub2. Cochrane Database Syst Rev. 2024. PMID: 39392114 Review. - Vestibular stimulation for promoting development and preventing morbidity in preterm infants.
Prescott MG, Wróblewska-Seniuk K, Lenells M, Fiander M, Soll R, Bruschettini M. Prescott MG, et al. Cochrane Database Syst Rev. 2024 Sep 5;9(9):CD016072. doi: 10.1002/14651858.CD016072. Cochrane Database Syst Rev. 2024. PMID: 39234929 Free PMC article. - Shorter versus longer duration antibiotic regimens for treatment of suspected neonatal sepsis.
Legge AA, Middleton JL, Fiander M, Cracknell J, Osborn DA, Gordon A. Legge AA, et al. Cochrane Database Syst Rev. 2024 Aug 30;8:CD016006. doi: 10.1002/14651858.CD016006. Cochrane Database Syst Rev. 2024. PMID: 39212160 - Testing of a risk-stratified patient decision aid to facilitate shared decision-making for extended postoperative thromboprophylaxis after major abdominal surgery for cancer.
Ivankovic V, Delisle M, Stacey D, Abou-Khalil J, Balaa F, Bertens KA, Dingley B, Martel G, McAlpine K, Nessim C, Tadros S, Carrier M, Auer RC. Ivankovic V, et al. Can J Surg. 2024 Aug 27;67(4):E320-E328. doi: 10.1503/cjs.014722. Print 2024 Jul-Aug. Can J Surg. 2024. PMID: 39191449 Free PMC article.
References
- Chalmers I, Enkin M, Keirse MJNC. Preparing and updating systematic reviews of randomized controlled trials of health care. Milbank Q. 1993;71(3):411‐437. - PubMed
- Lefebvre C, Manheimer E, Glanville J. Searching for studies In: Higgins J, Green S, eds. Cochrane Handbook for Systematic Reviews of Interventions. 5.1.0 ed. The Cochrane Collaboration; 2011.
- Joachims T. Text categorization with support vector machines: learning with many relevant features In: Nédellec C, Rouveirol C, eds. Machine Learning: ECML‐98. Vol.1398 Lecture Notes in Computer Science Berlin, Heidelberg: Springer Berlin Heidelberg; 1998.
- McCallum, Andrew , Nigam Kamal, and Others . 1998. “A comparison of event models for naive Bayes text classification.” In AAAI‐98 Workshop on Learning for Text Categorization, 752:41–48 Citeseer
- Goldberg, Yoav . 2015. “A primer on neural network models for natural language processing.” arXiv [cs.CL] . arXiv.http://arxiv.org/abs/1510.00726.
MeSH terms
Grants and funding
- MR/N015185/1/Medical Research Council (UK)
- R03 HS025024/HS/AHRQ HHS/United States
- R03-HS025024/Agency for Healthcare Research and Quality
- 5UH2CA203711-02,/U.S. National Library of Medicine
- Cochrane via Project Transform
- UH2 CA203711/CA/NCI NIH HHS/United States
- MR/N015185/1/MRC_/Medical Research Council/United Kingdom
- MR/J005037/1/MRC_/Medical Research Council/United Kingdom
- R01-LM012086-01A1/U.S. National Library of Medicine
- R01 LM012086/LM/NLM NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources