Cyclic di-adenosine monophosphate (c-di-AMP) is required for osmotic regulation in Staphylococcus aureus but dispensable for viability in anaerobic conditions - PubMed (original) (raw)
Cyclic di-adenosine monophosphate (c-di-AMP) is required for osmotic regulation in Staphylococcus aureus but dispensable for viability in anaerobic conditions
Merve S Zeden et al. J Biol Chem. 2018.
Abstract
Cyclic di-adenosine monophosphate (c-di-AMP) is a recently discovered signaling molecule important for the survival of Firmicutes, a large bacterial group that includes notable pathogens such as Staphylococcus aureus However, the exact role of this molecule has not been identified. dacA, the S. aureus gene encoding the diadenylate cyclase enzyme required for c-di-AMP production, cannot be deleted when bacterial cells are grown in rich medium, indicating that c-di-AMP is required for growth in this condition. Here, we report that an S. aureus dacA mutant can be generated in chemically defined medium. Consistent with previous findings, this mutant had a severe growth defect when cultured in rich medium. Using this growth defect in rich medium, we selected for suppressor strains with improved growth to identify c-di-AMP-requiring pathways. Mutations bypassing the essentiality of dacA were identified in alsT and opuD, encoding a predicted amino acid and osmolyte transporter, the latter of which we show here to be the main glycine betaine-uptake system in S. aureus. Inactivation of these transporters likely prevents the excessive osmolyte and amino acid accumulation in the cell, providing further evidence for a key role of c-di-AMP in osmotic regulation. Suppressor mutations were also obtained in hepS, hemB, ctaA, and qoxB, coding proteins required for respiration. Furthermore, we show that dacA is dispensable for growth in anaerobic conditions. Together, these findings reveal an essential role for the c-di-AMP signaling network in aerobic, but not anaerobic, respiration in S. aureus.
Keywords: Staphylococcus aureus (S. aureus); cyclic diadenosine monophosphate (c-di-AMP); osmotic swelling; respiration; signaling.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
Figure 1.
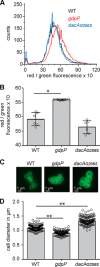
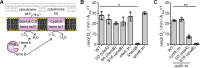
Variations in c-di-AMP levels impact the membrane potential and size of S. aureus cells. A and B, membrane potential measurement using a fluorescence-activated cell sorting-based method. WT LAC* as well as the high c-di-AMP gdpP and low c-di-AMP dacAG206S mutant strains were grown overnight in TSB medium. Cells were washed and mixed with DiOC2(3) and the green and red fluorescence intensities detected using a FACSCalibur cytometer. The fluorescence intensities of 10,000 gated events were recorded at the height of their emission peak. The ratio of red/green fluorescence × 10 was calculated for each event using the FlowJo V7 software, and representative histograms of cell counts versus fluorescence ratio are shown (A). B, mean values of red/green fluorescence × 10 were determined from the histograms in A, and the averages and standard deviations from six biological replicates were plotted. C and D, bacterial cell size determination by microscopy. WT, gdpP, and dacAG206S mutant strains were grown overnight in TSB medium. Culture aliquots were stained with vancomycin-BODIPY, and cells were imaged using a fluorescence microscope. C, representative images of WT, gdpP, and dacAG206S cells are shown. D, bacterial cell diameters were determined by drawing a line through the middle of non-dividing cells using ImageJ. 150 cells were measured (three biological replicates with 50 cells each), and the average cell diameters in micrometers and standard deviations were plotted. Statistical analysis was performed in Prism (GraphPad) using a Kruskal-Wallis test followed by a Dunn's multiple comparison test. Adjusted p values <0.05 are indicated by a single asterisk and adjusted p values < 0.01 by a double asterisk.
Figure 2.
The S. aureus dacA mutant can grow in CDM but not in TSB medium. A, bacterial growth on agar plates. WT LAC* and the isogenic dacA mutant were streaked on TSA or CDM plates, and images of plates were taken after overnight incubation at 37 °C. B, plating efficiencies. Bacterial suspensions were prepared for the WT and dacA mutant strains, and appropriate dilutions were spread on TSA or CDM plates and CFUs per ml of culture per _A_600 unit determined and plotted. The average values and standard deviations from four biological replicates were plotted.
Figure 3.
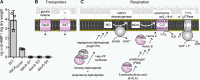
S. aureus dacA suppressor strains do not produce c-di-AMP but acquire mutations in genes coding for transport or respiration-related proteins. A, determination of cellular c-di-AMP levels by LC-MS/MS. Cell extracts (three biological replicates) were prepared from the suppressor strains _dacA-_S1, _dacA-_S2, and _dacA-_S4, and the production of c-di-AMP was assessed by LC-MS/MS. The extracts were prepared and analyzed at the same time as WT LAC* and the low-level c-di-AMP dacAG206S strain as reported in a previous study (20). No c-di-AMP could be detected in extracts derived from strains _dacA-_S1, _dacA-_S2, and _dacA-_S4, and c-di-AMP levels determined for the WT and dacAG206S mutant as part of the previous study (20) are shown as controls. B and C, schematic representation of proteins whose genes were found to be mutated in dacA suppressor strains and their predicted cellular functions. B, schematic representation and function of the OpuD and AlsT transporters. OpuD is a predicted (and as part of this study experimentally confirmed) glycine betaine and a reported weak proline transporter (43); AlsT is a predicted
l
-alanine/sodium symporter, but this substrate specificity could not be confirmed as part of this study. C, simplified view of the aerobic respiratory chain in S. aureus. The NADH dehydrogenase oxidizes NADH to NAD+, and electrons are transferred onto MQ to yield MQH2. The electrons are subsequently moved onto the heme A component of the cytochrome _aa_3 complex, composed of the QoxA, -B, -C, and -D proteins. Upon reduction of ½O2 to H2O by the cytochrome complex, protons (H+) are extruded and utilized by the F1F0-ATPase for the production of ATP. Suppressor mutations were identified in genes coding for HepS, HemB, CtaA (a membrane protein but shown for simplicity as a soluble protein), and QoxB, required for MQ, HemA, and cytochrome _aa_3 complex formation, respectively. The proteins for which gene variations were found in suppressor strains are indicated in purple in the schematics.
Figure 4.
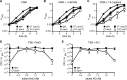
S. aureus dacA suppressors with mutations in opuD and alsT have improved growth in TSB. A and B, bacterial growth curves. WT LAC*, the dacA mutant, and suppressor strains S7 (opuD), S10 (alsT), and S18 (qoxB/Δ_opuD) (or short S18 (Δ_opuD)) were propagated in CDM (A) or TSB (B) medium, and their growth was monitored by taking A_600 readings. The average values and standard deviations from three independent experiments were plotted. C, genetic complementation of opuD mutants. Bacterial suspensions were prepared for WT, the dacA mutant, as well as suppressor strains S7 (opuD) and S18 (qoxB/Δ_opuD) (or short S18 (Δ_opuD_)) containing the empty piTET vector or the complementation plasmid piTET-opuD. Appropriate culture dilutions were plated on TSA plates containing 200 ng/ml Atet and CFUs per ml culture per A_600 unit determined and plotted. The average values and standard deviations from three experiments were plotted. D, opuD mutants are defective in glycine betaine uptake. WT, the dacA mutant, and the suppressor strains S7 (opuD) and S18 (qoxB/Δ_opuD) (or short S18 (Δ_opuD_)) containing the empty piTET vector or the plasmid piTET-opuD were grown to mid-log phase in CDM supplemented with 200 ng/ml Atet. For uptake assays, radiolabeled glycine betaine was added to culture aliquots; samples were removed and filtered at the indicated time points, and the radioactivity accumulated in the cells was measured. The average values and standard deviations from four experiments were plotted. E, alanine uptake assays. WT, dacA, and S10 (alsT) strains were grown to mid-log phase in CDM containing half the
l
-alanine concentration as in the standard medium. Bacterial suspensions were prepared, and radiolabeled alanine was added to the cultures. Sample aliquots were removed at the indicated time points and filtered, and the radioactivity accumulated in the cells was measured. The average values and standard deviations from four experiments were plotted.
Figure 5.
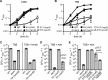
Glycine betaine and peptides inhibit and salt improves the growth of the dacA mutant. A–C, bacterial growth curves. WT LAC*, the dacA mutant, and S7 (opuD) and _dacA_-S10 (alsT) suppressor strains were propagated in CDM (A), CDM with 1 m
m
glycine betaine (GB) (B), or CDM with 1% tryptone (C) and their growth was monitored by determining _A_600 readings. The average values and standard deviations from three independent experiments were plotted. D and E, plating efficiencies of S. aureus on TSA plates supplemented with increasing amounts of NaCl or KCl. Bacterial suspensions were prepared for WT LAC*, and the dacA mutant strain and serial dilutions spotted on TSA or TSA plates containing the indicated concentrations of NaCl (D) or KCl (E). The average CFUs per ml culture per _A_600 unit and the standard deviations from three experiments were plotted.
Figure 6.
_S. aureus dacA s_uppressors with mutations in ctaA, hemB, or qoxB have improved growth in TSB that can be complemented chemically or genetically. A and B, bacterial growth curves. WT LAC*, the dacA mutant, and the suppressor strains S3 (ctaA), S4 (hemB), S13 (hepS), and S16 (qoxB) were propagated in CDM (A) or TSB (B) medium and their growth monitored by determining _A_600 readings. The average values and standard deviations of three independent experiments were plotted. C, chemical complementation analysis of the hemB mutant. Bacterial suspensions were prepared for WT LAC*, the dacA mutant, and suppressor strain S4 (hemB) and appropriate dilutions plated on TSA or TSA plates containing 10 μ
m
hemin. CFUs per ml culture per A_600 unit were determined and plotted. D and E, genetic complementation analysis of ctaA (D) and qoxB (E) mutants. Bacterial suspensions were prepared for WT LAC*, the dacA mutant, as well as suppressor strain S3 (ctaA) with or without the complementation plasmid piTET-ctaA (C) or suppressor strains S16 (qoxB) and S18 (qoxB/Δ_opuD) (D) with or without the complementation plasmid piTET_-qoxB._ Dilutions were plated on TSA plates containing 200 ng/ml Atet and the average CFUs per ml of culture per _A_600 unit and standard deviations from three experiments plotted.
Figure 7.
Oxygen consumption rates of WT, mutant, and dacA suppressor strains. A, schematic representation of the two S. aureus terminal oxidases. The main terminal oxidase Qox (also referred to as cytochrome _aa_3) is composed of the proteins QoxA–QoxD and has been suggested to contain heme A and heme B as cofactors (10) and requires the CtaA protein (a membrane protein but shown for simplicity as soluble protein) for heme A biosynthesis. The second terminal oxidase Cyd (also referred to as cytochrome bd) is composed of the proteins CydA and CydB and has been suggested to contain heme B and heme O (10) and therefore likely does not require CtaA for its synthesis. B and C, oxygen consumption rates of WT, mutant, and dacA suppressor strains. The indicated S. aureus strains were grown to mid-log phase, washed, and the oxygen consumption rates of concentrated culture aliquots determined following the addition of glucose. The oxygen consumption rates are plotted as nanomoles of O2 consumed per min per _A_600 = 1. For each strain, 3–6 biological replicates were used, and the average values and standard deviations of the oxygen consumption rates were plotted. Statistical analysis was performed in Prism (GraphPad) using a Kruskal-Wallis test followed by a Dunn's multiple comparison test. Adjusted p values <0.05 are indicated by a single asterisk and adjusted p values <0.01 by a double asterisk.
Figure 8.
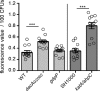
Reduced c-di-AMP levels lead to an increase in ROS production. Determination of endogenous ROS production in WT and mutant S. aureus strains is shown. LAC* (WT), the isogenic dacAG206S, and gdpP mutant strains as well as control strains SH1000 and SH1000Δ_katA_Δ_ahpC_ were grown to mid-log phase in TSB medium. Endogenous ROS production was determined using the indicator dye 2′,7′-dichlorofluorescein diacetate, which was used at a final concentration of 10 μ
m
. Fluorescence values were measured at excitation and emission wavelengths of 485 and 538 nm, respectively. All fluorescent values were normalized for CFUs, and the average fluorescence values and standard deviations per 100 CFUs from 12 biological replicates were plotted. Statistical analysis was performed in Prism (GraphPad) using for the comparison of the values obtained for the WT LAC* stain with those obtained for the isogenic dacAG206S and gdpP mutant strains (light gray bars) a Kruskal-Wallis test followed by a Dunn's multiple comparison test. For the comparison of the values obtained for strains SH1000 and SH1000Δ_katA_Δ_ahpC_ (medium gray bars), a Mann-Whitney test was used. Statistically significant differences are indicated by asterisks with p values <0.001 indicated by a triple asterisk.
Figure 9.
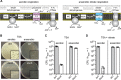
dacA is dispensable for the growth of S. aureus under anaerobic conditions. A, schematic overview of aerobic (left panel) versus anaerobic (right panel) respiration in S. aureus. Under aerobic conditions, oxygen is used as the terminal electron acceptor through the Qox system. In the absence of oxygen but in the presence of nitrate (NO3−), anaerobic respiration with nitrate as terminal electron acceptor through the nitrate reductase can occur. B, agar plate images. 10−4 dilutions of WT and dacA mutant cultures were plated on TSA plates, the plates incubated under aerobic or anaerobic conditions, and images taken after overnight incubation at 37 °C. Insets show representative magnified areas of the respective plate. C and D, plating efficiencies of WT and dacA mutant strains. Bacterial suspensions were plated on TSA (C) or TSA plates containing 20 m
m
KNO3 (D), and the plates were incubated aerobically or anaerobically as stated. Average CFUs per ml of culture per _A_600 unit and standard deviations from three experiments were plotted.
Figure 10.
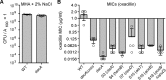
dacA mutant and suppressor strains have increased oxacillin sensitivity. A, plating efficiencies of WT and dacA mutant strains on Müller-Hinton agar plates supplemented with 2% NaCl. Bacterial suspensions were prepared and plated on Müller-Hinton agar plates supplemented with 2% NaCl, and CFUs per ml of culture per _A_600 units were determined. The average values and standard deviations from three experiments were plotted. B, oxacillin MICs for WT and mutant S. aureus strains. Bacterial suspensions were prepared for WT LAC*, the low-level c-di-AMP _dacA_G206S strain, the dacA mutant, and the indicated suppressor strains and spread on Müller-Hinton agar plates with 2% NaCl. Next, oxacillin MIC EvaluatorTM strips (Thermo Fisher Scientific) were placed on the plates, and the plates were incubated for 24 h at 35 °C. The average MIC values and standard deviations from four samples were plotted.
Similar articles
- Identification of the main glutamine and glutamate transporters in Staphylococcus aureus and their impact on c-di-AMP production.
Zeden MS, Kviatkovski I, Schuster CF, Thomas VC, Fey PD, Gründling A. Zeden MS, et al. Mol Microbiol. 2020 Jun;113(6):1085-1100. doi: 10.1111/mmi.14479. Epub 2020 Feb 11. Mol Microbiol. 2020. PMID: 31997474 Free PMC article. - New Insights into the Cyclic Di-adenosine Monophosphate (c-di-AMP) Degradation Pathway and the Requirement of the Cyclic Dinucleotide for Acid Stress Resistance in Staphylococcus aureus.
Bowman L, Zeden MS, Schuster CF, Kaever V, Gründling A. Bowman L, et al. J Biol Chem. 2016 Dec 30;291(53):26970-26986. doi: 10.1074/jbc.M116.747709. Epub 2016 Nov 10. J Biol Chem. 2016. PMID: 27834680 Free PMC article. - Inhibition of the Staphylococcus aureus c-di-AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM.
Tosi T, Hoshiga F, Millership C, Singh R, Eldrid C, Patin D, Mengin-Lecreulx D, Thalassinos K, Freemont P, Gründling A. Tosi T, et al. PLoS Pathog. 2019 Jan 22;15(1):e1007537. doi: 10.1371/journal.ppat.1007537. eCollection 2019 Jan. PLoS Pathog. 2019. PMID: 30668586 Free PMC article. - c-di-AMP: An Essential Molecule in the Signaling Pathways that Regulate the Viability and Virulence of Gram-Positive Bacteria.
Fahmi T, Port GC, Cho KH. Fahmi T, et al. Genes (Basel). 2017 Aug 7;8(8):197. doi: 10.3390/genes8080197. Genes (Basel). 2017. PMID: 28783096 Free PMC article. Review. - Cyclic di-AMP Signaling in Bacteria.
Stülke J, Krüger L. Stülke J, et al. Annu Rev Microbiol. 2020 Sep 8;74:159-179. doi: 10.1146/annurev-micro-020518-115943. Epub 2020 Jun 30. Annu Rev Microbiol. 2020. PMID: 32603625 Review.
Cited by
- An atypical GdpP enzyme linking cyclic nucleotide metabolism to osmotic tolerance and gene regulation in Mycoplasma bovis.
Zhu X, Baranowski E, Hao Z, Li X, Zhao G, Dong Y, Chen Y, Hu C, Chen H, Citti C, Wang A, Guo A. Zhu X, et al. Front Microbiol. 2023 Nov 30;14:1250368. doi: 10.3389/fmicb.2023.1250368. eCollection 2023. Front Microbiol. 2023. PMID: 38098652 Free PMC article. - Making and Breaking of an Essential Poison: the Cyclases and Phosphodiesterases That Produce and Degrade the Essential Second Messenger Cyclic di-AMP in Bacteria.
Commichau FM, Heidemann JL, Ficner R, Stülke J. Commichau FM, et al. J Bacteriol. 2018 Dec 7;201(1):e00462-18. doi: 10.1128/JB.00462-18. Print 2019 Jan 1. J Bacteriol. 2018. PMID: 30224435 Free PMC article. Review. - Two Ways To Convert a Low-Affinity Potassium Channel to High Affinity: Control of Bacillus subtilis KtrCD by Glutamate.
Krüger L, Herzberg C, Warneke R, Poehlein A, Stautz J, Weiß M, Daniel R, Hänelt I, Stülke J. Krüger L, et al. J Bacteriol. 2020 May 27;202(12):e00138-20. doi: 10.1128/JB.00138-20. Print 2020 May 27. J Bacteriol. 2020. PMID: 32253343 Free PMC article. - The Many Roles of the Bacterial Second Messenger Cyclic di-AMP in Adapting to Stress Cues.
Zarrella TM, Bai G. Zarrella TM, et al. J Bacteriol. 2020 Dec 7;203(1):e00348-20. doi: 10.1128/JB.00348-20. Print 2020 Dec 7. J Bacteriol. 2020. PMID: 32839175 Free PMC article. Review. - mSphere of Influence: Metabolic redundancies enhance pathogenesis.
Lehman MK. Lehman MK. mSphere. 2024 Jul 30;9(7):e0023924. doi: 10.1128/msphere.00239-24. Epub 2024 Jul 3. mSphere. 2024. PMID: 38958458 Free PMC article.
References
- Fridkin S. K., Hageman J. C., Morrison M., Sanza L. T., Como-Sabetti K., Jernigan J. A., Harriman K., Harrison L. H., Lynfield R., Farley M. M., and Active Bacterial Core Surveillance Program of the Emerging Infections Program Network (2005) Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352, 1436–1444 10.1056/NEJMoa043252 - DOI - PubMed
- Francis J. S., Doherty M. C., Lopatin U., Johnston C. P., Sinha G., Ross T., Cai M., Hansel N. N., Perl T., Ticehurst J. R., Carroll K., Thomas D. L., Nuermberger E., and Bartlett J. G. (2005) Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin. Infect. Dis. 40, 100–107 10.1086/427148 - DOI - PubMed
- Pagels M., Fuchs S., Pané-Farré J., Kohler C., Menschner L., Hecker M., McNamarra P. J., Bauer M. C., von Wachenfeldt C., Liebeke M., Lalk M., Sander G., von Eiff C., Proctor R. A., and Engelmann S. (2010) Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol. Microbiol. 76, 1142–1161 10.1111/j.1365-2958.2010.07105.x - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 100289/WT_/Wellcome Trust/United Kingdom
- 260371/ERC_/European Research Council/International
- BB/L024209/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/P028225/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases