Oxidation of SQSTM1/p62 mediates the link between redox state and protein homeostasis - PubMed (original) (raw)
doi: 10.1038/s41467-017-02746-z.
Elsje G Otten 1, Diego Manni 1, Rhoda Stefanatos 1, Fiona M Menzies 2, Graham R Smith 1 3, Diana Jurk 1, Niall Kenneth 1, Simon Wilkinson 4, Joao F Passos 1, Johannes Attems 5, Elizabeth A Veal 1, Elisa Teyssou 6, Danielle Seilhean 6 7, Stéphanie Millecamps 6, Eeva-Liisa Eskelinen 8, Agnieszka K Bronowska 9, David C Rubinsztein 2 10, Alberto Sanz 1, Viktor I Korolchuk 11
Affiliations
- PMID: 29343728
- PMCID: PMC5772351
- DOI: 10.1038/s41467-017-02746-z
Oxidation of SQSTM1/p62 mediates the link between redox state and protein homeostasis
Bernadette Carroll et al. Nat Commun. 2018.
Abstract
Cellular homoeostatic pathways such as macroautophagy (hereinafter autophagy) are regulated by basic mechanisms that are conserved throughout the eukaryotic kingdom. However, it remains poorly understood how these mechanisms further evolved in higher organisms. Here we describe a modification in the autophagy pathway in vertebrates, which promotes its activity in response to oxidative stress. We have identified two oxidation-sensitive cysteine residues in a prototypic autophagy receptor SQSTM1/p62, which allow activation of pro-survival autophagy in stress conditions. The Drosophila p62 homologue, Ref(2)P, lacks these oxidation-sensitive cysteine residues and their introduction into the protein increases protein turnover and stress resistance of flies, whereas perturbation of p62 oxidation in humans may result in age-related pathology. We propose that the redox-sensitivity of p62 may have evolved in vertebrates as a mechanism that allows activation of autophagy in response to oxidative stress to maintain cellular homoeostasis and increase cell survival.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Fig. 1
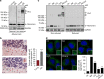
p62 forms oligomers and aggregates in response to oxidation. a Mouse brain tissue, young (3 months) and old (24 months) analysed by immunoblotting for p62, PRDX-SO3 and actin as a loading control in reducing (2.5% β-ME) and non-reducing conditions. DLC disulphide-linked conjugates. Arrows indicate the positions of monomeric and oligomeric p62. b Representative images and quantification of p62 aggregates in old mouse Purkinje cells in the cerebellum. Green arrows and red arrowheads indicate Purkinje cells positive and negative for p62 aggregates, respectively. c Effect of autophagy inhibition (bafilomycin A1 (Baf, 400 nM, 4 h) and chloroquine (CQ, 50 μM, 4 h)) and oxidative stress (H2O2 (3 mM, 10 min) and PR-619 (5 μM, 30 min)) on p62 DLC (c) and p62 aggregation (d) in HeLa cells. d Anti-p62 staining analysed by confocal microscopy. Error bars represent s.e.m., n = 3, *P < 0.05, **P < 0.01, ***P < 0.005 (unpaired _t_-tests). Asterisk indicates a non-specific band. Scale bar: 20 µm
Fig. 2
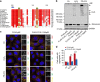
Two conserved cysteine residues located in a disordered region of p62 are required for formation of DLC and ubiquitylated aggregates. a Alignment showing cysteines 105 and 113 (highlighted) are conserved in vertebrates. Increasing conservation across species is shown by light-to-dark red. b p62 −/− MEFs stably expressing either wild type or C105A,C113A FLAG-p62 were treated with H2O2 (3 mM, 1 min) or PR-619 (5 μM, 10 min) and immunoblotted in non-reducing conditions. c Formation of ubiquitylated, p62-positive aggregates in stable cells described in b following oxidative stress (1 mM H2O2 or 5 μM PR-619 for 30 min). Cells were immunostained for ubiquitin and p62, analysed by confocal microscopy (c) and quantified (d). Error bars represent s.e.m., n = 3, **P < 0.01, ***P < 0.005 (unpaired _t_-tests). Arrows indicate the positions of monomeric and oligomeric p62. Scale bar: 10 µm
Fig. 3
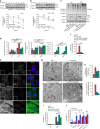
Oxidation-sensitive p62 is required for pro-survival autophagy. a p62 −/− MEFs stably expressing FLAG-tagged wild type or C105A,C113A p62 were treated with cycloheximide (CHX, 50 μg/ml) and H2O2 (1 mM), either in the absence (a) or presence (b) of bafilomycin A1 (Baf, 50 nM), lysed at the indicated time post treatment, immunoblotted for p62 and quantified. c Cells described in a plus one stably expressing K7A,D69A PB1-domain mutant of p62 were treated with H2O2 (1 mM, 5 h), lysed and immunoblotted for ubiquitin, LC3, p62 and actin (c) and quantified (d). e, f Stable p62 cell lines in control conditions were fixed and stained for p62 and LC3 (e) and the % cells with >20 autophagosomes was quantified (f). g, h Electron microscopy of stable p62 cell lines. White arrows: autophagosomes; black arrows: autolysosomes; blue arrow: endosome. Scale bar: 500 nm (g). Autophagosomes were quantified and graphs represent average number and % cells with three or more autophagosomes per field of view (h). i, j Stable p62 cell lines were treated as in c and % cell death was analysed by Ready Probes fluorescent dyes (i). j Stable p62 cell lines were treated as indicated and % cell death was analysed as in i. Error bars represent s.e.m., n = 3, *P < 0.05, **P < 0.01, ***P < 0.005 (unpaired _t_-tests). NS not significant. Scale bar: 20 µm (IF) and 500 nm (TEM)
Fig. 4
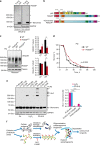
Oxidation-sensitive p62 is important for the oxidative stress resistance of flies and is perturbed in human age-related disease. a Whole-fly lysates were analysed in reducing (2.5% β-ME) and non-reducing conditions for p62 homologue, Ref(2)P. Asterisk indicates a non-specific band. b Diagram representing the introduction of an 18 amino acid fragment of human p62 containing C105 and C113 to produce a ‘humanised’ Ref(2)P (Ref(2)Pox) in flies using CRISPR/Cas9. c Wild-type (WT) and Ref(2)Pox flies were treated with paraquat (PQ, 20 mM) for 12 h and whole-fly lysates were analysed by immunoblot for ubiquitin, Ref(2)P and tubulin and quantified. Error bars represent s.e.m., n = 3 (at least 10 flies per group per replicate); *P < 0.05 (unpaired _t_-test). d Combined survival data (of three repeats) of wild-type versus Ref(2)Pox flies in the presence of 20 mM PQ. Survival was assessed every 12 h. At least 60 flies per group per replicate were used and log-rank statistics applied. e _p62_−/− MEFs stably expressing FLAG-tagged wild-type p62, C105A,C113A p62 or ALS-associated mutation K102E p62 were treated with H2O2 (500 μM, 1 min) and PR-619 (20 μM, 10 min), analysed for p62 DLC formation in non-reducing conditions and quantified. Error bars represent s.e.m., n = 3; ***P < 0.005 (unpaired _t_-tests). f Diagram of the proposed role for p62 oxidation in the aggregation and degradation of autophagy substrates. Formation of p62 DLC is triggered by oxidative stress, which promotes degradation of p62 and bound substrates (e.g. ubiquitylated proteins) through autophagy. Mutations in p62 sequence (e.g. ALS-related K102E) can impair the formation of DLC
Comment in
- Oxidation of p62 as an evolutionary adaptation to promote autophagy in stress conditions.
Otten EG, Stefanatos R, Carroll B, Korolchuk VI. Otten EG, et al. Cell Stress. 2018 Mar 23;2(4):91-93. doi: 10.15698/cst2018.04.132. Cell Stress. 2018. PMID: 31225472 Free PMC article.
Similar articles
- The selective autophagy receptor SQSTM1/p62 improves lifespan and proteostasis in an evolutionarily conserved manner.
Aparicio R, Hansen M, Walker DW, Kumsta C. Aparicio R, et al. Autophagy. 2020 Apr;16(4):772-774. doi: 10.1080/15548627.2020.1725404. Epub 2020 Feb 10. Autophagy. 2020. PMID: 32041473 Free PMC article. - p62/SQSTM1/Sequestosome-1 is an N-recognin of the N-end rule pathway which modulates autophagosome biogenesis.
Cha-Molstad H, Yu JE, Feng Z, Lee SH, Kim JG, Yang P, Han B, Sung KW, Yoo YD, Hwang J, McGuire T, Shim SM, Song HD, Ganipisetti S, Wang N, Jang JM, Lee MJ, Kim SJ, Lee KH, Hong JT, Ciechanover A, Mook-Jung I, Kim KP, Xie XQ, Kwon YT, Kim BY. Cha-Molstad H, et al. Nat Commun. 2017 Jul 24;8(1):102. doi: 10.1038/s41467-017-00085-7. Nat Commun. 2017. PMID: 28740232 Free PMC article. - p62/Sequestosome-1, Autophagy-related Gene 8, and Autophagy in Drosophila Are Regulated by Nuclear Factor Erythroid 2-related Factor 2 (NRF2), Independent of Transcription Factor TFEB.
Jain A, Rusten TE, Katheder N, Elvenes J, Bruun JA, Sjøttem E, Lamark T, Johansen T. Jain A, et al. J Biol Chem. 2015 Jun 12;290(24):14945-62. doi: 10.1074/jbc.M115.656116. Epub 2015 Apr 30. J Biol Chem. 2015. PMID: 25931115 Free PMC article. - Histone H3F3/H3.3 chaperone DAXX converts to modulate SQSTM1 phase condensation for NFE2L2 activation.
Yang Y, Valionyte E, Kelly J, Luo S. Yang Y, et al. Autophagy. 2020 Jan;16(1):171-172. doi: 10.1080/15548627.2019.1677323. Epub 2019 Oct 17. Autophagy. 2020. PMID: 31607206 Free PMC article. Review. - p62/SQSTM1 and Selective Autophagy in Cardiometabolic Diseases.
Jeong SJ, Zhang X, Rodriguez-Velez A, Evans TD, Razani B. Jeong SJ, et al. Antioxid Redox Signal. 2019 Aug 20;31(6):458-471. doi: 10.1089/ars.2018.7649. Epub 2019 Feb 11. Antioxid Redox Signal. 2019. PMID: 30588824 Free PMC article. Review.
Cited by
- AKR1C1 connects autophagy and oxidative stress by interacting with SQSTM1 in a catalytic-independent manner.
Chang LL, Li YK, Zhao CX, Zeng CM, Ge FJ, Du JM, Zhang WZ, Lu PH, He QJ, Zhu H, Yang B. Chang LL, et al. Acta Pharmacol Sin. 2022 Mar;43(3):703-711. doi: 10.1038/s41401-021-00673-w. Epub 2021 May 20. Acta Pharmacol Sin. 2022. PMID: 34017066 Free PMC article. - Naked Mole-Rat Cortex Maintains Reactive Oxygen Species Homeostasis During In Vitro Hypoxia or Ischemia and Reperfusion.
Eaton L, Wang T, Roy M, Pamenter ME. Eaton L, et al. Curr Neuropharmacol. 2023;21(6):1450-1461. doi: 10.2174/1570159X20666220327220929. Curr Neuropharmacol. 2023. PMID: 35339183 Free PMC article. - Ubr1-induced selective endophagy/autophagy protects against the endosomal and Ca2+-induced proteostasis disease stress.
Wang BB, Xu H, Isenmann S, Huang C, Elorza-Vidal X, Rychkov GY, Estévez R, Schittenhelm RB, Lukacs GL, Apaja PM. Wang BB, et al. Cell Mol Life Sci. 2022 Mar 1;79(3):167. doi: 10.1007/s00018-022-04191-8. Cell Mol Life Sci. 2022. PMID: 35233680 Free PMC article. - Use the Protonmotive Force: Mitochondrial Uncoupling and Reactive Oxygen Species.
Berry BJ, Trewin AJ, Amitrano AM, Kim M, Wojtovich AP. Berry BJ, et al. J Mol Biol. 2018 Oct 19;430(21):3873-3891. doi: 10.1016/j.jmb.2018.03.025. Epub 2018 Apr 4. J Mol Biol. 2018. PMID: 29626541 Free PMC article. Review. - Enhanced Levels of Peroxisome-Derived H2O2 Do Not Induce Pexophagy but Impair Autophagic Flux in HEK-293 and HeLa Cells.
Li H, Lismont C, Costa CF, Hussein MAF, Baes M, Fransen M. Li H, et al. Antioxidants (Basel). 2023 Mar 2;12(3):613. doi: 10.3390/antiox12030613. Antioxidants (Basel). 2023. PMID: 36978861 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- BB/H022384/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/L016354/1/MRC_/Medical Research Council/United Kingdom
- G1100540/MRC_/Medical Research Council/United Kingdom
- G0900652/MRC_/Medical Research Council/United Kingdom
- 12825/CRUK_/Cancer Research UK/United Kingdom
- G0400074/MRC_/Medical Research Council/United Kingdom
- G0502157/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases