Epitranscriptomic m6A Regulation of Axon Regeneration in the Adult Mammalian Nervous System - PubMed (original) (raw)
. 2018 Jan 17;97(2):313-325.e6.
doi: 10.1016/j.neuron.2017.12.036.
Xu Wang 2, Ran An 3, Jessica Cassin 4, Caroline Vissers 5, Yuanyuan Liu 6, Yajing Liu 7, Tianlei Xu 8, Xinyuan Wang 9, Samuel Zheng Hao Wong 10, Jessica Joseph 11, Louis C Dore 12, Qiang Dong 13, Wei Zheng 14, Peng Jin 15, Hao Wu 8, Bin Shen 6, Xiaoxi Zhuang 16, Chuan He 12, Kai Liu 2, Hongjun Song 17, Guo-Li Ming 18
Affiliations
- PMID: 29346752
- PMCID: PMC5777326
- DOI: 10.1016/j.neuron.2017.12.036
Epitranscriptomic m6A Regulation of Axon Regeneration in the Adult Mammalian Nervous System
Yi-Lan Weng et al. Neuron. 2018.
Abstract
N6-methyladenosine (m6A) affects multiple aspects of mRNA metabolism and regulates developmental transitions by promoting mRNA decay. Little is known about the role of m6A in the adult mammalian nervous system. Here we report that sciatic nerve lesion elevates levels of m6A-tagged transcripts encoding many regeneration-associated genes and protein translation machinery components in the adult mouse dorsal root ganglion (DRG). Single-base resolution m6A-CLIP mapping further reveals a dynamic m6A landscape in the adult DRG upon injury. Loss of either m6A methyltransferase complex component Mettl14 or m6A-binding protein Ythdf1 globally attenuates injury-induced protein translation in adult DRGs and reduces functional axon regeneration in the peripheral nervous system in vivo. Furthermore, Pten deletion-induced axon regeneration of retinal ganglion neurons in the adult central nervous system is attenuated upon Mettl14 knockdown. Our study reveals a critical epitranscriptomic mechanism in promoting injury-induced protein synthesis and axon regeneration in the adult mammalian nervous system.
Keywords: CNS axon regeneration; DRG; Mettl14; PNS axon regeneration; RGC; YTHDF1; epitranscriptomics; mRNA methylation; protein synthesis.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests.
Figures
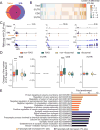
Figure 1. SNL upregulates levels of m6A-tagged mRNAs encoding RAGs and protein translation machinery in adult DRGs in vivo
(A) Venn diagram of m6A-tagged transcripts identified by m6A-SMART-seq in adult mouse DRGs under naïve and SNL D1 conditions. (B) Venn diagram of all m6A-tagged genes at SNL D1 and known RAGs. (C) Scatter plot of expression levels of m6A-tagged transcripts under naïve and SNL D1 conditions. Lines indicate 2 fold differences and RAGs are indicated by magenta dots. (D) Heatmap diagrams of the m6A transcript levels under naïve and SNL D1 conditions for a select group of RAGs and genes related to protein translation functions. (E) m6A-MeRIP Q-PCR validation of differential m6A transcript levels under naïve and SNL D1 conditions for selected RAGs. Values are normalized to the naïve condition and represent mean ± SEM (n = 3 experimental replications from 6 animals; *P < 0.05; **P < 0.01; t-test). (F) GO enrichment analyses of the top 400 genes with increased m6A-tagged transcript levels (orange) and the top 400 genes with decreased m6A-tagged transcript levels (black) at SNL D1. (G) Scatter plots of log2 fold changes of m6A-tagged and total transcript levels between naïve and SNL D1 conditions. Subsets of genes are labeled with different colors in the same plot: RAGs (magenta), ribosomal subunit-related genes (red), translation initiation-related genes (blue), and translation regulation-related genes (yellow). See also Figure S1.
Figure 2. SNL modifies the m6A landscape of transcriptomes of adult mouse DRGs in vivo
(A) Venn diagram of m6A tagged-transcripts identified by m6A-CLIP-SMART-seq in adult mouse DRGs under naïve and SNL D1 conditions. (B) Dynamic changes of m6A sites in transcripts from adult DRGs at SNL D1. Cn8ghanges of m6A sites are plotted for the whole transcript (total) and in different sub-transcript regions (5′ UTR, CDS, and 3′ UTR). CDS: coding sequence region. (C) m6A-CLIP-SMART-seq examples for multiple RAGs. Shown are sample tracks for both m6A-CLIP-seq (top panels) and RNA-seq (bottom panels). CLIP unique tag coverage is shown in black, and m6A sites are indicated with vertical lines. (D) Comparison of dynamic m6A sites between RAGs and non-RAGs, and between transcripts encoding ribosomal subunit-related and non-ribosomal subunit-related proteins, in different transcript regions (total, 5′ UTR, CDS, and 3′ UTR) under naïve and SNL D1 conditions. Values represent mean differential m6A tag numbers (n = 154 RAGs and 5,867 non RAGs; n = 55 ribosomal subunit-related and 5,966 non-ribosomal subunit-related genes; ***P < 0.001; one-way ANOVA with Tukey’s post hoc test). (E) GO enrichment analyses of transcripts with differential m6A sites at SNL D1. See also Figure S2.
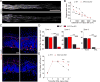
Figure 3. Mettl14 deletion attenuates SNL-induced global protein translation and ATF3 protein expression in the adult DRG
(A) m6A dot-blot showing diminished m6A levels in mRNA from DRGs of adult Syn-Cre;Mettl14 cKO mice. Methylene blue was used to assess the equal loading of mRNA. Representative images (top panel) and quantification (bottom panel) are shown. Values represent mean ± SEM (n = 2 animals per group; ***P < 0.001; two-way ANOVA). (**B**) Box plots depicting the fold changes of the gene expression level between RAGs and non-RAGs after injury in WT and Mettl14_cKO DRGs. Each box shows the first quartile, median, and third quartile (***P < 0.001;#_P_ > 0.05; one-way ANOVA with Tukey’s post hoc test). (C-D) SUnSET analysis of new protein synthesis in adult L4/5 DRGs of WT and Mettl14 cKO mice. De novo synthesized proteins were pulse-chase labeled for one hour after injection of puromycin at SNL D1. Western blot of DRG lysates were performed for different conditions. GAPDH was used as the loading control. Representative images (C) and quantification (D) are shown. Values are normalized to the WT naïve condition and plots represent ranges of mean ± SEM (n = 4 animals; **P < 0.01; *P < 0.05; two-way ANOVA). See Figure S3E for images from different exposures of the same Western blot example. (E-F) Assessment of ATF3 induction in WT and_Mettl14 cKO DRGs at SNL D1. Sample images of ATF3 immunostaining (E) and quantification (F) are shown. Scale bars: 50 μm. Values represent mean ± SEM (n = 4 animals; ***P < 0.001; two-way ANOVA). (G-H) Time-course analysis of ATF3 induction in WT and_Mettl14_ cKO adult DRGs. Immunoassay of DRG protein lysates were performed by capillary electrophoresis. GAPDH was used as the loading control. Sample images of blots (G) and quantification (H) are shown. Values represent mean ± SEM (n = 3 animals; **P < 0.01; two-way ANOVA). See also Figure S3.
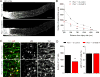
Figure 4. Mettl14 deletion attenuates functional axon regeneration of adult DRG neurons in vivo
(A-B) Analysis of regeneration of sensory axons by SCG10 immunostaining at SNL D3 in adult WT and _Mettl14f/f_mice upon intrathecal injection of AAV2/9 to express Cre. Sample images of regenerating sensory axons identified by SCG10 (A; scale bar: 1 mm) and quantification (B) are shown. SCG10 immunofluorescence intensity was measured at different distal distances and normalized to that at 1 mm before the lesion site as the regenerative index. Values represent mean ± SEM (n = 8 animals for WT and 10 animals for AAV-Cre;Mettl14 cKO; ***P < 0.001; **P < 0.01; two-way ANOVA). (C-D) Assay for re-innervation of the hindpaw epidermal area by regenerating sensory axons. Sample images of cross sections of hindpaw glabrous skin of WT and AAV-Cre;Mettl14 cKO mice immunostained with the pan neuronal marker PGP9.5 are shown (C). The dotted line indicates the border between dermis and epidermis. Scale bar: 20 μm. Also shown are quantifications of the number of intra-epidermal nerve fibers in a 1 mm segment of different epidermal areas (D). Values represent mean ± SEM (n = 5 animals per group; ***P < 0.001; **P < 0.01; two-way ANOVA). (E) Assessment of thermal sensory recovery after SNL in WT and AAV-Cre;Mettl14 cKO mice. Values represent mean ± SEM (n = 10 animals per group; ***P < 0.001; two-way ANOVA). See also Figure S4.
Figure 5. YTHDF1 is required for injury-induced global de novo protein synthesis and robust axon regeneration of adult DRG neurons
(A-B) SUnSET analysis of new protein synthesis in adult L4/5 DRGs of WT and Ythdf1 KO mice. De novo synthesized proteins were pulse-chase labeled for one hour after injection of puromycin at SNL D1. Western blot of DRG lysates were performed for different conditions. GAPDH was used as the loading control. Representative images (A) and quantification (B) are shown. Values are normalized to WT naïve conditions and plots represent ranges of mean ± SEM (n = 3 for WT and Ythdf1 KO each; ***P < 0.01; **P < 0.01; two-way ANOVA). See Figure S5C for images from different exposures of the same Western blot example. (C-D) Analysis of regeneration of sensory axons by SCG10 immunostaining at SNL D3 in adult WT and Ythdf1 KO mice. Sample images of regenerating sensory axons identified by SCG10 (C; scale bar: 1 mm) and quantification (D) are shown. SCG10 immunofluorescence intensity was measured at different distal distances and normalized to the level 1 mm before the lesion site as the regenerative index. Values represent mean ± SEM (n = 7 animals for WT and 6 animals for Ythdf1 KO mice; ***P < 0.001; **P < 0.01; two-way ANOVA). See also Figure S5.
Figure 6. Mettl14 is required for robust Pten deletion-induced axonal regeneration of retinal ganglion neurons in the adult mouse CNS
Adult Ptenf/f mice were co-injected with AAV-Cre and AAV-shRNA-control or AAV-shRNA-Mettl14. Optic nerve was crushed 4 weeks after AAV injection and RGC axons were traced by fluorescence conjugated cholera toxin B (FITC-CTB) 2 weeks later. Shown are sample images of sections of optic nerve containing FITC-CTB-labeled axons (A; scale bar: 200 μm) and quantification of numbers of regenerating axons at different distances from the injury site (B). Values represent mean + SEM (n = 5 animals per group; ** P < 0.01; *P < 0.05; ANOVA followed by Fisher’s LSD). Also shown are sample images of whole-mount retina with Tuj1 (green) and pS6 (red) immunostaining (C; scale bar: 50 μm) and quantification of densities of Tuj1+ RGCs and percentages of Tuj1+ RGCs expressing pS6. Values represent mean + SEM (n = 5 animals per each group; **P < 0.01; Student’s t test). See also Figure S6.
Comment in
- Neural repair: Tagging mRNA drives regeneration.
Whalley K. Whalley K. Nat Rev Neurosci. 2018 Mar;19(3):121. doi: 10.1038/nrn.2018.14. Epub 2018 Feb 8. Nat Rev Neurosci. 2018. PMID: 29416127 No abstract available.
Similar articles
- Modest enhancement of sensory axon regeneration in the sciatic nerve with conditional co-deletion of PTEN and SOCS3 in the dorsal root ganglia of adult mice.
Gallaher ZR, Steward O. Gallaher ZR, et al. Exp Neurol. 2018 May;303:120-133. doi: 10.1016/j.expneurol.2018.02.012. Epub 2018 Feb 16. Exp Neurol. 2018. PMID: 29458059 Free PMC article. - Promoting axon regeneration by inhibiting RNA N6-methyladenosine demethylase ALKBH5.
Wang D, Zheng T, Zhou S, Liu M, Liu Y, Gu X, Mao S, Yu B. Wang D, et al. Elife. 2023 Aug 3;12:e85309. doi: 10.7554/eLife.85309. Elife. 2023. PMID: 37535403 Free PMC article. - The Unfolded Protein Response and Cholesterol Biosynthesis Link Luman/CREB3 to Regenerative Axon Growth in Sensory Neurons.
Ying Z, Zhai R, McLean NA, Johnston JM, Misra V, Verge VM. Ying Z, et al. J Neurosci. 2015 Oct 28;35(43):14557-70. doi: 10.1523/JNEUROSCI.0012-15.2015. J Neurosci. 2015. PMID: 26511246 Free PMC article. - Lab review: Molecular dissection of the signal transduction pathways associated with PTEN deletion-induced optic nerve regeneration.
Huang H, Kaur S, Hu Y. Huang H, et al. Restor Neurol Neurosci. 2019;37(6):545-552. doi: 10.3233/RNN-190949. Restor Neurol Neurosci. 2019. PMID: 31839616 Free PMC article. Review. - Epigenetic and epitranscriptomic regulation of axon regeneration.
Cheng Y, Song H, Ming GL, Weng YL. Cheng Y, et al. Mol Psychiatry. 2023 Apr;28(4):1440-1450. doi: 10.1038/s41380-023-02028-9. Epub 2023 Mar 15. Mol Psychiatry. 2023. PMID: 36922674 Free PMC article. Review.
Cited by
- Cell type-specific regulation of m6 A modified RNAs in the aging Drosophila brain.
Perlegos AE, Byrns CN, Bonini NM. Perlegos AE, et al. Aging Cell. 2024 Mar;23(3):e14076. doi: 10.1111/acel.14076. Epub 2024 Jan 11. Aging Cell. 2024. PMID: 38205931 Free PMC article. - Identification and validation of m6A RNA regulatory network in pulpitis.
Xu H, Chen G, Zhou J, Zhou X, Wang P, Chen C, Xu Z, Lv F, Li X. Xu H, et al. BMC Oral Health. 2023 Nov 17;23(1):878. doi: 10.1186/s12903-023-03578-8. BMC Oral Health. 2023. PMID: 37978362 Free PMC article. - The rise of epitranscriptomics: recent developments and future directions.
Cerneckis J, Ming GL, Song H, He C, Shi Y. Cerneckis J, et al. Trends Pharmacol Sci. 2024 Jan;45(1):24-38. doi: 10.1016/j.tips.2023.11.002. Epub 2023 Dec 15. Trends Pharmacol Sci. 2024. PMID: 38103979 Free PMC article. Review. - Comprehensive Analysis of Differential m6A RNA Methylomes in the Hippocampus of Cocaine-Conditioned Mice.
Xue A, Huang Y, Li M, Wei Q, Bu Q. Xue A, et al. Mol Neurobiol. 2021 Aug;58(8):3759-3768. doi: 10.1007/s12035-021-02363-4. Epub 2021 Apr 7. Mol Neurobiol. 2021. PMID: 33826069 - N6-Methyladenosine Role in Acute Myeloid Leukaemia.
Ianniello Z, Fatica A. Ianniello Z, et al. Int J Mol Sci. 2018 Aug 9;19(8):2345. doi: 10.3390/ijms19082345. Int J Mol Sci. 2018. PMID: 30096915 Free PMC article. Review.
References
- Befort K, Karchewski L, Lanoue C, Woolf CJ. Selective up-regulation of the growth arrest DNA damage-inducible gene Gadd45 alpha in sensory and motor neurons after peripheral nerve injury. The European journal of neuroscience. 2003;18:911–922. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RM1 HG008935/HG/NHGRI NIH HHS/United States
- T32 GM007445/GM/NIGMS NIH HHS/United States
- R37 NS047344/NS/NINDS NIH HHS/United States
- R01 DA043361/DA/NIDA NIH HHS/United States
- P01 NS097206/NS/NINDS NIH HHS/United States
- R35 NS097370/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous