Interactions, localization, and phosphorylation of the m6A generating METTL3-METTL14-WTAP complex - PubMed (original) (raw)
Interactions, localization, and phosphorylation of the m6A generating METTL3-METTL14-WTAP complex
Eva Schöller et al. RNA. 2018 Apr.
Abstract
_N_6-methyladenine (m6A) is found on many eukaryotic RNAs including mRNAs. m6A modification has been implicated in mRNA stability and turnover, localization, or translation efficiency. A heterodimeric enzyme complex composed of METTL3 and METTL14 generates m6A on mRNAs. METTL3/14 is found in the nucleus where it is localized to nuclear speckles and the splicing regulator WTAP is required for this distinct nuclear localization pattern. Although recent crystal structures revealed how the catalytic MT-A70 domains of METTL3 and METTL14 interact with each other, a more global architecture including WTAP and RNA interactions has not been reported so far. Here, we used recombinant proteins and mapped binding surfaces within the METTL3/14-WTAP complex. Furthermore, we identify nuclear localization signals and identify phosphorylation sites on the endogenous proteins. Using an in vitro methylation assay, we confirm that monomeric METTL3 is soluble and inactive while the catalytic center of METTL14 is degenerated and thus also inactive. In addition, we show that the C-terminal RGG repeats of METTL14 are required for METTL3/14 activity by contributing to RNA substrate binding. Our biochemical work identifies characteristic features of METTL3/14-WTAP and reveals novel insight into the overall architecture of this important enzyme complex.
Keywords: METTL14; METTL3; RNA modification; m6A; methyltransferase.
© 2018 Schöller et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures
FIGURE 1.
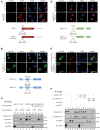
Binding studies of METTL3, METTL14, and WTAP. (A) Coomassie gel of purified recombinant full-length GST-METTL3 and copurified METTL14. The coexpression was conducted in insect cells (SF21). The complex was purified via the GST-tag on METTL3. (B) Schematic representation of the different truncation constructs of F/H-METTL3 and myc-METTL14 used to narrow down the binding sites of these proteins. (C) Coomassie gel of recombinantly expressed and purified complexes composed of the indicated truncated METTL14 and METTL3 (76–580) constructs. Complexes were purified via GST-METTL3 (76–580) and GST was subsequently cleaved off during elution. (D) Schematic view of the interaction surface between METTL3 and METTL14 revealed by coexpression studies. (E) Scheme of the different WTAP N- and C-terminal truncation constructs. (F) Western blots of coimmunoprecipitations of full-length F/H-METTL3 with N-terminal truncations of myc-WTAP. The upper panel was incubated with α-HA antibody, the lower one with α-myc antibody. The arrows show the bands for the different constructs. (G) Western blots of coimmunoprecipitations as in F but with C-terminally truncated constructs of myc-WTAP. (H) Western blot of coimmunoprecipitated full-length myc-WTAP and LH-deletion mutants of F/H-METTL3 (ΔLH-METTL3). (I) Western blots of coimmunoprecipitations of the N-terminal part of F/H-METTL3 (leader helix, short LH) with C-terminally truncated constructs of myc-WTAP. The upper panel shows the HA-antibody treated blot, the lower blot shows the α-myc-western blot. (J) Schematic cartoon of the regions of interaction between WTAP and METTL3 based on the results of coimmunoprecipitation experiments.
FIGURE 2.
Localization studies of METTL3, METTL14, and WTAP. (A) Immunofluorescence staining of HeLa cells transfected with wild-type (WT) myc-WTAP (upper panels) and myc-WTAP-NLS mutant (mut) (lower panels). The schematic cartoon under the immunofluorescences shows the very N-terminal location and the sequence of the NLS in the protein. (B) Immunofluorescence of HeLa cells which were transfected with WT (upper panels) and NLS-mutated F/H-METTL3 (lower panels). The computed NLS is predicted in the potential RBD between the leader helix and the MT-A70 domain as shown in the schematic view beneath the stainings. (C) Western blots of coimmunoprecipitations between F/H-METTL3-NLS mutant and myc-WTAP. GFP containing either a myc- or a F/H-tag serves as control. (D) HeLa cells were transfected with myc-METTL14 WT (upper panels) and the predicted F/H-NLS mutant (lower panels). The cartoon shows the position and sequence of the potential NLS in the protein. (E) For the upper panel, HEK293T cells were transfected with myc-METTL14 and F/H-METTL3. The lower panels show immunofluorescence stainings of myc-METTL14 and the predicted F/H-METTL3-NLS mutant. (F) Western blot of the coimmunoprecipitation of different F/H-METTL3 and myc-METTL14 constructs. GFP was used as control.
FIGURE 3.
Establishment of a monoclonal antibody against METTL3. (A) The upper western blot shows IPs of F/H-METTL3 using an α-FLAG antibody for precipitation on the one hand and the established α-METTL3 antibody 29C8 on the other. The blot was incubated with an α-HA antibody. The lower panel shows a western blot of an endogenous METTL3-IP using the METTL3 antibody. (B) Coomassie gel of endogenous METTL3 purified using the anti-METTL3 antibody 29C8. METTL14 is coimmunoprecipitated. The control IP does not show any bands.
FIGURE 4.
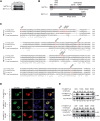
Identification and characterization of phosphorylation sites in METTL3/14 complex. (A) Coimmunoprecipitation of METTL3 and METTL14. To analyze phosphorylation sites, endogenous METTL3 was purified from nuclear HeLa S3 lysate by α-METTL3 29C8 antibody. Endogenous METTL14 was coimmunoprecipitated. (B) Mass spectrometric measurements of human METTL3/14 complex. Schematic representation of phosphorylation sites in METTL3 and METTL14 proteins. The structural domains (leader helix [LH]; nuclear localization sequence [NLS]; N-terminal α-helical motif [NHM]; C-terminal motif [CTM]) are shown in gray and phosphorylation sites are represented by red bars. (C) Conservation of the phosphorylation sites detected in METTL3 and METTL14. Phosphorylation sites of METTL3 and METTL14 measured in our analysis are shown in red. (D) Nuclear localization of METTL3 S219 phosphorylation variants. F/H-METTL3 constructs were detected by α-FLAG antibody staining (green). Myc-METTL14 was visualized by α-myc antibody (red). Nuclei were stained with DAPI (blue). (E) Impact of the LH-surrounding phosphorylation sites on WTAP interaction. HEK293T cells were transfected with F/H-METTL3 phosphorylation mutants together with myc-WTAP. Complexes were purified by α-FLAG-IPs. F/H-METTL3 constructs and copurified myc-WTAP were subsequently visualized by western blotting.
FIGURE 5.
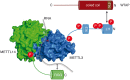
Phosphorylation sites of METTL3 and METTL14 are not essential for methylation activity. (A) Phosphorylation sites in the central region of METTL3 do not influence the interaction with myc-METTL14. HEK293T cells were transfected with F/H-METTL3 phosphorylation mutants together with myc-METTL14. Complexes were purified by α-FLAG-IPs. F/H-METTL3 constructs and copurified myc-METTL14 were visualized by western blotting. (B) Measurement of methyltransferase activity of METTL3/14 wild-type and mutated variants. Point mutations in the catalytic center of both proteins were introduced as indicated in the schematic representation. Data are shown as mean ± SD from six independent replicates. As control, 1 µg of recombinant protein was separated on a 10% SDS gel and stained with Coomassie blue. (C) Methyltransferase activity of WT METTL3/14 complex and variants in the phosphorylation residues T348 and S350. Data are shown as mean ± SD from five independent replicates. As control, 1 µg of recombinant protein was separated on a 10% SDS gel and stained with Coomassie blue. (D) Structural environment of phosphorylated residue S399 in METTL14 (green) relating to R471 in METTL3 (blue). (Left) pS399 position within the METTL3/14 interaction surface. (Right) Potential contact of pS399 to the R471 residue. (E) Coimmunoprecipitation of F/H-METTL3 with S399A and S399E mutants of myc-METTL14. HEK293T cells were transfected with F/H-METTL3 and myc-METTL14 constructs. F/H-METTL3 was purified by α-FLAG-IP. All myc-METTL14 constructs were copurified and visualized by western blotting. (F) In vitro methyltransferase activity of WT METTL3/14 and mutants in the phosphorylated S399 residue of METTL14. Phosphorylation residues S399 of METTL14 was modified as shown in the schematic representation. Data are shown as mean ± SD from six independent replicates. For control, 1 µg of recombinant protein was separated on a 10% SDS gel and stained with Coomassie blue. Of note, primary antibodies against the HA- and the myc-tags were added simultaneously to the same western blot membrane. Since F/H-METTL3 and myc-METTL14 migrate identically on the gel, western blots appear very similar (A,E).
FIGURE 6.
RGG deletion affects catalytic activity of METTL3/14 and substrate binding (A) Analysis of methyltransferase activity of METTL3 and METTL14 lacking the RGG repeats. Truncations were introduced to the METTL14 sequence (METTL14 ΔRGG) directly or mediated by TEV protease cleavage (METTL14 TEV-RGG) as indicated in the schematic representation. Data are shown as mean ± SD from five independent replicates. As control, 1 µg of recombinant protein was separated on a SDS gel and stained with Coomassie blue. (B) RNA binding of full-length METTL3/14 and METTL3/14 ΔRGG. The proteins were diluted serially in a 50 µL reaction mixture. (C) RNA-binding studies of F/H-METTL14 using cross-linking and immunoprecipitation (CLIP). F/H-METTL3 with F/H-METTL14-WT or F/H-METTL14-ΔRGG were transfected into HEK293T cells grown in 4SU-containg medium and UV cross-linked (356 nm). After α-FLAG-IP and RNase T1 digestion, the bound RNA was radioactively labeled, the protein-RNA complex was separated by SDS-PAGE and blotted onto a nitrocellulose membrane. Cross-linked RNA was visualized by autoradiography.
FIGURE 7.
Schematic representation of the interaction surfaces of METTL3 (blue), METTL14 (green), and WTAP (red). Proposed working model includes phosphorylation sites (red) of METTL3/14 as well as the NLS motifs of METTL3 (blue) and WTAP (brown). METTL3 catalyzes methylation of the adenosine base (red) within the RRACH motif. METTL14 coordinates and stabilizes the RNA binding.
Similar articles
- METTL3 regulates m6A in endometrioid epithelial ovarian cancer independently of METTl14 and WTAP.
Ma Z, Li Q, Liu P, Dong W, Zuo Y. Ma Z, et al. Cell Biol Int. 2020 Dec;44(12):2524-2531. doi: 10.1002/cbin.11459. Epub 2020 Sep 11. Cell Biol Int. 2020. PMID: 32869897 - Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase.
Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, Zhao X, Li A, Yang Y, Dahal U, Lou XM, Liu X, Huang J, Yuan WP, Zhu XF, Cheng T, Zhao YL, Wang X, Rendtlew Danielsen JM, Liu F, Yang YG. Ping XL, et al. Cell Res. 2014 Feb;24(2):177-89. doi: 10.1038/cr.2014.3. Epub 2014 Jan 10. Cell Res. 2014. PMID: 24407421 Free PMC article. - Dynamic assembly of the mRNA m6A methyltransferase complex is regulated by METTL3 phase separation.
Han D, Longhini AP, Zhang X, Hoang V, Wilson MZ, Kosik KS. Han D, et al. PLoS Biol. 2022 Feb 10;20(2):e3001535. doi: 10.1371/journal.pbio.3001535. eCollection 2022 Feb. PLoS Biol. 2022. PMID: 35143475 Free PMC article. - Human m6A writers: Two subunits, 2 roles.
Wang X, Huang J, Zou T, Yin P. Wang X, et al. RNA Biol. 2017 Mar 4;14(3):300-304. doi: 10.1080/15476286.2017.1282025. Epub 2017 Jan 25. RNA Biol. 2017. PMID: 28121234 Free PMC article. Review. - Role of WTAP in Cancer: From Mechanisms to the Therapeutic Potential.
Fan Y, Li X, Sun H, Gao Z, Zhu Z, Yuan K. Fan Y, et al. Biomolecules. 2022 Sep 2;12(9):1224. doi: 10.3390/biom12091224. Biomolecules. 2022. PMID: 36139062 Free PMC article. Review.
Cited by
- The Regulation of RNA Modification Systems: The Next Frontier in Epitranscriptomics?
Schaefer MR. Schaefer MR. Genes (Basel). 2021 Feb 26;12(3):345. doi: 10.3390/genes12030345. Genes (Basel). 2021. PMID: 33652758 Free PMC article. Review. - Cryo-EM structures of human m6A writer complexes.
Su S, Li S, Deng T, Gao M, Yin Y, Wu B, Peng C, Liu J, Ma J, Zhang K. Su S, et al. Cell Res. 2022 Nov;32(11):982-994. doi: 10.1038/s41422-022-00725-8. Epub 2022 Sep 27. Cell Res. 2022. PMID: 36167981 Free PMC article. - RNA modifications in aging-associated cardiovascular diseases.
Yang X, Gokulnath P, Lehmann HI, Hou Z, Yang S, You L, Zhang G, Xing Y, Lei J, Li G, Guo S, Shang H. Yang X, et al. Aging (Albany NY). 2022 Sep 29;14(19):8110-8136. doi: 10.18632/aging.204311. Epub 2022 Sep 29. Aging (Albany NY). 2022. PMID: 36178367 Free PMC article. Review. - Functions of RNA N6-methyladenosine modification in acute myeloid leukemia.
Zheng X, Gong Y. Zheng X, et al. Biomark Res. 2021 May 17;9(1):36. doi: 10.1186/s40364-021-00293-w. Biomark Res. 2021. PMID: 34001273 Free PMC article. Review. - RNA-binding proteins in cardiovascular biology and disease: the beat goes on.
Völkers M, Preiss T, Hentze MW. Völkers M, et al. Nat Rev Cardiol. 2024 Jun;21(6):361-378. doi: 10.1038/s41569-023-00958-z. Epub 2024 Jan 2. Nat Rev Cardiol. 2024. PMID: 38163813 Review.
References
- Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46: 674–690. - PubMed
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. - PubMed
- Fu Y, Dominissini D, Rechavi G, He C. 2014. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet 15: 293–306. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases