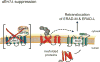The Dfm1 Derlin Is Required for ERAD Retrotranslocation of Integral Membrane Proteins - PubMed (original) (raw)
The Dfm1 Derlin Is Required for ERAD Retrotranslocation of Integral Membrane Proteins
Sonya Neal et al. Mol Cell. 2018.
Erratum in
- The Dfm1 Derlin Is Required for ERAD Retrotranslocation of Integral Membrane Proteins.
Neal S, Jaeger PA, Duttke SH, Benner C, Glass CK, Ideker T, Hampton RY. Neal S, et al. Mol Cell. 2018 Mar 1;69(5):915. doi: 10.1016/j.molcel.2018.02.014. Mol Cell. 2018. PMID: 29499140 No abstract available.
Abstract
Endoplasmic reticulum (ER)-associated degradation (ERAD) removes misfolded proteins from the ER membrane and lumen by the ubiquitin-proteasome pathway. Retrotranslocation of ubiquitinated substrates to the cytosol is a universal feature of ERAD that requires the Cdc48 AAA-ATPase. Despite intense efforts, the mechanism of ER exit, particularly for integral membrane (ERAD-M) substrates, has remained unclear. Using a self-ubiquitinating substrate (SUS), which undergoes normal retrotranslocation independently of known ERAD factors, and the new SPOCK (single plate orf compendium kit) micro-library to query all yeast genes, we found the rhomboid derlin Dfm1 was required for retrotranslocation of both HRD and DOA ERAD pathway integral membrane substrates. Dfm1 recruited Cdc48 to the ER membrane with its unique SHP motifs, and it catalyzed substrate extraction through its conserved rhomboid motifs. Surprisingly, dfm1Δ can undergo rapid suppression, restoring wild-type ERAD-M. This unexpected suppression explained earlier studies ruling out Dfm1, and it revealed an ancillary ERAD-M retrotranslocation pathway requiring Hrd1.
Keywords: Cdc48; DOA; Dfm1; ER; ERAD; HMG-CoA reductase; HRD; derlins; endoplasmic reticulum; retrotranslocation; rhomboid.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS: The authors declare they have no competing interests within the contents of this article.
Figures
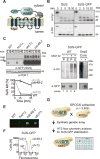
Figure 1. SUS-GFP behaves as a HRD pathway substrate and is used as an optical retrotranslocation factor reporter in the yeast genomic screen
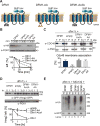
(A) Depiction of fusion protein, SUS-GFP. The transmembrane Hmg1 domain has a lumenal Myc epitope and the cytosolic domain has three HA epitopes followed by the HRD RING domain fused with the GFP epitope. (B) SUS-GFP is correctly inserted into microsomes. Microsomes prepared from strains expressing SUS or SUS-GFP were digested with trypsin for the indicated times and immunoblotted with α-Myc and α-HA. (C) Degradation of SUS-GFP depends on Ubc7 and cdc48-2. The indicated strains expressing SUS-GFP were grown into log phase and degradation was measured by a cycloheximide chase (CHX). After CHX addition, cells were lysed at the indicated times and analyzed by SDS-PAGE and immunoblotted for SUS-GFP with α-GFP. Band intensities were normalized to PGK1 loading control and quantified by ImageJ. t=0 was taken as 100% and data is represented as mean ± SEM from at least three experiments. (D) Full-length SUS-GFP retrotranslocates in vivo. Left panel, in vivo retrotranslocation of SUS-GFP. WT strains were grown to log-phase and treated with MG132 (25 μg/mL). Crude lysate was prepared and ultracentrifuged to discern ubiquitinated SUS-GFP that either has been retrotranslocated into the soluble fraction (S) or remained in the membrane (P). Following fractionation, SUS-GFP was immunoprecipitated from both fractions, resolved on 8% SDS-PAGE and immunoblotted for ubiquitin and SUS-GFP. Right panel, in vivo retrotranslocated SUS-GFP is full-length. Full-length SUS-GFP was immunoprecipitated and immunoblotted for SUS-GFP with α-GFP and α-Ubi. (E) Colony fluorescence shows increased steady-state levels of SUS-GFP in _ubc7_Δ and cdc48-2 strains. The indicated strains were grown to log phase and .03 OD of cells were spotted onto YPD plates and grown at 30°C for two days. Plates were imaged by a fluorescent imager. (F) Flow cytometry shows increased steady-state levels of SUS-GFP in _ubc7_Δ and cdc48-2 strains. The indicated strains were grown to log phase and were subjected to flow cytometry. Histograms of 10,000 cells are shown, with the number cells versus GFP fluorescence. (G) High throughput pipeline for identifying genes involved in SUS-GFP retrotranslocation. The SUS-GFP reporter was introduced into the SPOCK collection consisting of a 5,808 genome-wide library of yeast nulls and DaMP essential genes using SGA technology. The array was transferred to liquid YPD media in 384-well plates and SUS-GFP fluorescence was measured by a LS Fortessa high throughput flow cytometer.
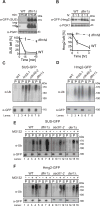
Figure 2. Dfm1 is required for in vivo Hmg2-GFP retrotranslocation
(A) Dfm1 is involved in degradation of SUS-GFP. WT and _dfm1_Δ strains were grown to log phase and degradation was measured by CHX. After CHX addition, cells were lysed at the indicated times and analyzed by SDS-PAGE and immunoblotted for SUS-GFP with α-GFP. (B) Dfm1 is involved in degradation of Hmg2-GFP. Same as (A), except CHX-chase assay was performed on Hmg2-GFP. (A–B) Band intensities were normalized to PGK1 loading control and quantified by ImageJ. t=0 was taken as 100% and data is represented as mean ± SEM from at least three experiments. (C) Dfm1 is required for retrotranslocation of SUS-GFP. Crude lysate was prepared from each strain and ultracentrifuged to discern ubiquitinated SUS-GFP that either has been retrotranslocated into the soluble fraction (S) or remained in the membrane (P). Following fractionation, SUS-GFP was immunoprecipitated from both fractions, resolved on 8% SDS-PAGE and immunoblotted with α-GFP and α-Ubi. (D) Dfm1 is required for retrotranslocation of Hmg2-GFP. Same as (C), except in vivo retrotranslocation assay was performed on Hmg2-GFP. (E–F) Der1 and Sec61 are not involved in retrotranslocation of SUS-GFP and Hmg2-GFP. Same as (C), except der1Δ and sec61-2 strains were grown to log-phase and treated with vehicle or MG132 prior to in vivo retrotranslocation assay.
Figure 3. Dfm1 is involved in degradation of integral membrane substrates
(A–C) Degradation of the indicated tagged ERAD-M and ERAD-C substrates were measured by CHX in isogenic strains. After CHX addition, cells were lysed at the indicated times, analyzed by SDS-PAGE and immunoblotted for each substrate. (D) Dfm1 is involved in retrotranslocation of Ste6*. The indicated strains were grown to log-phase and treated with MG132 (25 μg/mL). Crude lysate was prepared and ultracentrifuged to discern ubiquitinated Ste6*-GFP that either has been retrotranslocated into the soluble fraction (S) or remained in the membrane (P). Following fractionation, Ste6*-GFP was immunoprecipitated from both fractions, resolved on 8% SDS-PAGE and immunoblotted for ubiquitin and Ste6*-GFP. (E–G) Dfm1 is not involved in degradation of ERAD-L substrates. Same as (A–C), except CHX-chase assay was performed on the indicated ERAD-L substrates. (H) Dfm1 is involved in degradation of HRD1. Same as above, except CHX-chase assay was performed on Hrd1. (I) Dfm1 is involved in retrotranslocation of Hrd1 in vivo. Crude lysate was prepared from each strain and ultracentrifuged to discern ubiquitinated Hrd1-5xmyc that either has been retrotranslocated into the soluble fraction (S) or remained in the membrane fraction (P). Following fractionation, Hrd1-5xmyc was immunoprecipitated from both fractions, resolved on 8% SDS-PAGE and immunoblotted with α-Myc and α-Ubi. (A–C, E, & H) Band intensities were normalized to PGK1 loading control and quantified by ImageJ. t=0 was taken as 100% and data is represented as mean ± SEM from at least three experiments.
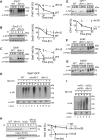
Figure 4. Dfm1 SHP box is required, but not sufficient for retrotranslocation
(A) Depiction of Dfm1, Der1, Dfm1-5Ashp, and Der1-Shp. Dfm1 and Der1 is an ER-localized membrane protein with six transmembrane domains (Greenblatt et al., 2011). Unlike Der1, Dfm1 has an extended cytoplasmic tail containing two SHP boxes. (B) Dfm1 is the major component for Cdc48 binding to the ER. Total cell lysate (T) from the indicated strains were separated into soluble cytosolic fraction (S) and pellet microsomal fraction (P) upon centrifugation at 14,000 × g. Each fraction was analyzed by SDS-PAGE and immunoblotted for Cdc48 with α-CDC48 and PGK1 with α-PGK1. (C) Stability of Dfm1 variants. Degradation of Dfm1, Dfm1-5Ashp, and Der1-Shp were measured by CHX-chase assay at the indicated times, cells were lysed, analyzed by SDS-PAGE and immunoblotted with α-HA. (D) SHP box is sufficient for Cdc48 recruitment. Same as (B), except strains expressing variants of Dfm1 were used. (E) Dfm1’s SHP box is required for degradation of Hmg2-GFP. In the indicated strains, degradation of Hmg2-GFP was measured by CHX-chase assay. Cells were analyzed by SDS-PAGE and immunoblotted for Hmg2-GFP with α-GFP. (F) in vivo retrotranslocation assay of Hmg2-GFP. After treatment with MG132, crude lysate was prepared from each strain and ultracentrifuged to discern ubiquitinated Hmg2-GFP that either has been retrotranslocated into the soluble fraction (S) or remained in the membrane (P). Following fractionation, Hmg2-GFP was immunoprecipitated from both fractions, resolved on 8% SDS-PAGE and immunoblotted with α-GFP and α-Ubi. (B, D) The graph shows the quantification of Cdc48 in the pellet fractions of the respective cells as measured from ImageJ. Data is represented as percentage of Cdc48 that is bound to pellet fraction and is shown as mean ± SEM from three independent experiments. (C, E) Band intensities were normalized to PGK1 loading control and quantified by ImageJ. t=0 was taken as 100% and data is represented as mean ± SEM from at least three experiments.
Figure 5. WR and GxxxG motif is required for Hmg2-GFP retrotranslocation
(A) Depiction of Dfm1, Dfm1-AA and Dfm1- Ax3A. Dfm1 has WR motif in the first lumenal loop and a GxxxG dimerization motif in the TMD. (B) Stability of Dfm1 variants. Degradation of Dfm1, Dfm1-AA, and Dfm1-Ax3A were measured by CHX-chase assay. At the indicated times, cells were lysed, analyzed by SDS-PAGE and immunoblotted with α-HA. (C) Dfm1-AA and Dfm1-Ax3A recruit Cdc48 to microsomes. Cell lysate (T) from the indicated strains were separated into soluble cytosolic fraction (S) and pellet microsomal fraction (P) upon centrifugation at 14,000 × g. Each fraction was analyzed by SDS-PAGE and immunoblotted for Cdc48 with α-CDC48 and PGK1 with α-PGK1. The graph shows the quantification of Cdc48 in the pellet fractions of the respective cells as measured from ImageJ. Data is represented as percentage of Cdc48 that is bound to pellet fraction and is shown as mean ± SEM from three independent experiments. (D) Dfm1’s WR and GxxxG motif is required for degradation of Hmg2-GFP. In the indicated strains, degradation of Hmg2-GFP was measured by CHX-chase assay. Cells were analyzed by SDS-PAGE and immunoblotted for Hmg2-GFP with α-GFP. (E) in vivo retrotranslocation assay of Hmg2-GFP. After treatment with MG132, crude lysate was prepared from each strain and ultracentrifuged to discern ubiquitinated Hmg2-GFP that either has been retrotranslocated into the soluble fraction (S) or remained in the membrane (P). Following fractionation, Hmg2-GFP was immunoprecipitated from both fractions, resolved on 8% SDS-PAGE and immunoblotted with α-GFP and α-Ubi. (B, D) Band intensities were normalized to PGK1 loading control and quantified by ImageJ. t=0 was taken as 100% and data is represented as mean ± SEM from at least three experiments.
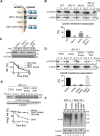
Figure 6. _dfm1_Δ rapidly suppresses overtime
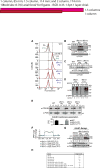
(A) dfm1Δ cells containing overexpressed SUS-GFP were passaged to suppression. The indicated cells with overexpressed SUS-GFP were passaged at the indicated number of times into fresh minimal media (P0, P4 and P11) and SUS-GFP levels were analyzed by flow cytometry. Histograms of 10,000 cells are shown, with the number of cells versus GFP fluorescence. Note: panels are aligned so all fluorescent histograms are comparable between panels (B) SUS-GFP is degraded to WT levels in _dfm1_Δ suppressed cells. Degradation of SUS-GFP was measured by CHX-chase assay in WT, _dfm1_Δ P0 and _dfm1_Δ P11 cells. After CHX addition, cells were lysed at the indicated times, analyzed by SDS-PAGE and immunoblotted for SUS-GFP with α-GFP. (C) Hmg2-GFP is degraded to WT levels in _dfm1_Δ suppressed cells. Same as (B) except degradation of Hmg2-GFP was measured by CHX-chase assay in WT, _dfm1_Δ P0 and _dfm1_Δ P9 cells. (D) in vivo Hmg2-GFP retrotranslocation completely restored _dfm1_Δ suppressed cells. Crude lysate was prepared from the indicated strains treated with vehicle or MG132 (25 μg/mL). Lysates were ultracentrifuged to discern ubiquitinated Hmg2-GFP that either has been retrotranslocated into the soluble fraction (S) or remained in the membrane (P). Following fractionation, Hmg2-GFP was immunoprecipitated from both fractions, resolved on 8% SDS-PAGE and immunoblotted with α-GFP and α-Ubi. (E) Cdc48 recruitment to microsomes is restored in _dfm1_Δ suppressed cells. Total cell lysate (T) from the indicated strains were separated into soluble cytosolic fraction (S) and pellet microsomal fraction (P) upon centrifugation at 14,000 × g. Each fraction was analyzed by SDS-PAGE and immunoblotted for Cdc48 with α-CDC48 and PGK1 with α-PGK1. The graph shows the quantification of Cdc48 in the pellet fractions of the respective cells as measured from ImageJ. Data is represented as percentage of Cdc48 that is bound to pellet fraction and is shown as mean ± SEM from three independent experiments. (F) ChrXV duplication is substrate induced upon loss of Dfm1. Chromosome profiles of whole genome sequencing data mapped across ChrXV. Genomic levels through entire ChrXV are twice as high in suppressed dfm1Δ cells expressing Hmg2-GFP or SUS-GFP with respect to _dfm1_Δ cells containing empty vector. (G) Hrd1 levels are upregulated in dfm1Δ suppressees. Degradation of Hrd1-5xmyc was measured by CHX-chase assay in _dfm1_Δ P0 and _dfm1_Δ P11 overexpressing Hmg2-GFP. After CHX addition, cells were lysed at the indicated times, analyzed by SDS-PAGE and immunoblotted for Hrd1 with α-Myc and Hmg2 with α-GFP. (H) The indicated strains overexpressing SUS-GFP were passaged to suppression.
Figure 7. Model of Hrd1-mediated suppression in the dfm1Δ null
_dfm1_Δ null mutants undergo rapid suppression when ERAD-M substrates are strongly expressed, revealing an ancillary Dfm1-independent retrotranslocation route for ERAD-M substrates, which mediated by Hrd1.
Comment in
- Membrane Protein Dislocation by the Rhomboid Pseudoprotease Dfm1: No Pore Needed?
Avci D, Lemberg MK. Avci D, et al. Mol Cell. 2018 Jan 18;69(2):161-162. doi: 10.1016/j.molcel.2017.12.031. Mol Cell. 2018. PMID: 29351840
Similar articles
- Inner-nuclear-membrane-associated degradation employs Dfm1-independent retrotranslocation and alleviates misfolded transmembrane-protein toxicity.
Flagg MP, Wangeline MA, Holland SR, Duttke SH, Benner C, Neal S, Hampton RY. Flagg MP, et al. Mol Biol Cell. 2021 Apr 1;32(7):521-537. doi: 10.1091/mbc.E20-11-0720. Epub 2021 Feb 10. Mol Biol Cell. 2021. PMID: 33566711 Free PMC article. - Derlin rhomboid pseudoproteases employ substrate engagement and lipid distortion to enable the retrotranslocation of ERAD membrane substrates.
Nejatfard A, Wauer N, Bhaduri S, Conn A, Gourkanti S, Singh N, Kuo T, Kandel R, Amaro RE, Neal SE. Nejatfard A, et al. Cell Rep. 2021 Oct 19;37(3):109840. doi: 10.1016/j.celrep.2021.109840. Cell Rep. 2021. PMID: 34686332 Free PMC article. - A Cdc48 "Retrochaperone" Function Is Required for the Solubility of Retrotranslocated, Integral Membrane Endoplasmic Reticulum-associated Degradation (ERAD-M) Substrates.
Neal S, Mak R, Bennett EJ, Hampton R. Neal S, et al. J Biol Chem. 2017 Feb 24;292(8):3112-3128. doi: 10.1074/jbc.M116.770610. Epub 2017 Jan 11. J Biol Chem. 2017. PMID: 28077573 Free PMC article. - Protein Quality Control of the Endoplasmic Reticulum and Ubiquitin-Proteasome-Triggered Degradation of Aberrant Proteins: Yeast Pioneers the Path.
Berner N, Reutter KR, Wolf DH. Berner N, et al. Annu Rev Biochem. 2018 Jun 20;87:751-782. doi: 10.1146/annurev-biochem-062917-012749. Epub 2018 Feb 2. Annu Rev Biochem. 2018. PMID: 29394096 Review. - The evolving role of ubiquitin modification in endoplasmic reticulum-associated degradation.
Preston GM, Brodsky JL. Preston GM, et al. Biochem J. 2017 Feb 15;474(4):445-469. doi: 10.1042/BCJ20160582. Biochem J. 2017. PMID: 28159894 Free PMC article. Review.
Cited by
- Membrane Protein Dimerization in Cell-Derived Lipid Membranes Measured by FRET with MC Simulations.
Škerle J, Humpolíčková J, Johnson N, Rampírová P, Poláchová E, Fliegl M, Dohnálek J, Suchánková A, Jakubec D, Strisovsky K. Škerle J, et al. Biophys J. 2020 Apr 21;118(8):1861-1875. doi: 10.1016/j.bpj.2020.03.011. Epub 2020 Mar 29. Biophys J. 2020. PMID: 32246901 Free PMC article. - Assays for studying normal versus suppressive ERAD-associated retrotranslocation pathways in yeast.
Bhaduri S, Neal SE. Bhaduri S, et al. STAR Protoc. 2021 Jul 7;2(3):100640. doi: 10.1016/j.xpro.2021.100640. eCollection 2021 Sep 17. STAR Protoc. 2021. PMID: 34278330 Free PMC article. - Potential Physiological Relevance of ERAD to the Biosynthesis of GPI-Anchored Proteins in Yeast.
Nakatsukasa K. Nakatsukasa K. Int J Mol Sci. 2021 Jan 21;22(3):1061. doi: 10.3390/ijms22031061. Int J Mol Sci. 2021. PMID: 33494405 Free PMC article. Review. - Inner-nuclear-membrane-associated degradation employs Dfm1-independent retrotranslocation and alleviates misfolded transmembrane-protein toxicity.
Flagg MP, Wangeline MA, Holland SR, Duttke SH, Benner C, Neal S, Hampton RY. Flagg MP, et al. Mol Biol Cell. 2021 Apr 1;32(7):521-537. doi: 10.1091/mbc.E20-11-0720. Epub 2021 Feb 10. Mol Biol Cell. 2021. PMID: 33566711 Free PMC article. - Accumulated precursors of specific GPI-anchored proteins upregulate GPI biosynthesis with ARV1.
Liu YS, Wang Y, Zhou X, Zhang L, Yang G, Gao XD, Murakami Y, Fujita M, Kinoshita T. Liu YS, et al. J Cell Biol. 2023 May 1;222(5):e202208159. doi: 10.1083/jcb.202208159. Epub 2023 Feb 24. J Cell Biol. 2023. PMID: 36828365 Free PMC article.
References
- Avci D, Fuchs S, Schrul B, Fukumori A, Breker M, Frumkin I, Chen C, Biniossek M, Kremmer E, Schilling O, et al. The Yeast ER-Intramembrane Protease Ypf1 Refines Nutrient Sensing by Regulating Transporter Abundance. Mol Cell. 2014;56:630–640. - PubMed
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM111024/GM/NIGMS NIH HHS/United States
- R37 DK051996/DK/NIDDK NIH HHS/United States
- R01 DK051996/DK/NIDDK NIH HHS/United States
- R35 GM133565/GM/NIGMS NIH HHS/United States
- R01 ES014811/ES/NIEHS NIH HHS/United States
- R41 TR001908/TR/NCATS NIH HHS/United States
- R01 GM084279/GM/NIGMS NIH HHS/United States
- R21 HL088083/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous