'Cold shock' increases the frequency of homology directed repair gene editing in induced pluripotent stem cells - PubMed (original) (raw)
'Cold shock' increases the frequency of homology directed repair gene editing in induced pluripotent stem cells
Q Guo et al. Sci Rep. 2018.
Abstract
Using CRISPR/Cas9 delivered as a RNA modality in conjunction with a lipid specifically formulated for large RNA molecules, we demonstrate that homology directed repair (HDR) rates between 20-40% can be achieved in induced pluripotent stem cells (iPSC). Furthermore, low HDR rates (between 1-20%) can be enhanced two- to ten-fold in both iPSCs and HEK293 cells by 'cold shocking' cells at 32 °C for 24-48 hours following transfection. This method can also increases the proportion of loci that have undergone complete sequence conversion across the donor sequence, or 'perfect HDR', as opposed to partial sequence conversion where nucleotides more distal to the CRISPR cut site are less efficiently incorporated ('partial HDR'). We demonstrate that the structure of the single-stranded DNA oligo donor can influence the fidelity of HDR, with oligos symmetric with respect to the CRISPR cleavage site and complementary to the target strand being more efficient at directing 'perfect HDR' compared to asymmetric non-target strand complementary oligos. Our protocol represents an efficient method for making CRISPR-mediated, specific DNA sequence changes within the genome that will facilitate the rapid generation of genetic models of human disease in iPSCs as well as other genome engineered cell lines.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Figure 1
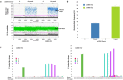
Single-stranded oligonucleotide (ssODN) donor design, droplet digital PCR probes and primer designs for gene editing and mutation detection at CAMK2D locus. (a) Two guide RNAs (CAMK-CR1 and CAMK-CR2) were designed to specifically target CAMK2D Exon2, CAMK-CR1 and CAMK-CR2 overlap by 14 nucleotides and were designed to cleave the DNA to introduce the same sequence alteration by HDR. (b) Two ssODN HDR donors (C-CR2 and C-CR2-Asym) were designed to introduce a kinase dead K43R mutation (AAA to AGG) and four silent mutations into Exon2 of the CAMK2D locus. The ssODN donor C-CR2 is a (+) strand HDR donor which is complementary to the guide RNA targeted cleavage strand with balanced homology arms around each side of the intended mutations (5′-73nt and 3′-72nt, respectively). C-CR2-Asym is a (−) strand HDR donor which is complementary to the guide RNA non-targeted strand with homology arms that differ in length (5′-93nt and 3′-36nt, respectively). To prevent subsequent re-cleavage, both donor oligo C-CR2 and C-CR2-Asym introduces three silent mutations within the guide CAMK-CR1 recognition site and one silent mutation within the PAM site. C-CR2 and C-CR2-Asym introduces four silent mutations within the guide CAMK-CR2 recognition site. A pair of primers and allele-specific probes conjugated with Vic or Fam fluorophores were also designed to detect separately the unaltered wild type alleles and mutated sequence conversion events. The forward primer was designed to anneal within the donor sequence while the reverse primer was designed to anneal outside of the donor sequence to ensure the proper locus was amplified.
Figure 2
An optimized method for co-delivery of a single-stranded oligonucleotide (ssODN) donor and sgRNA/Cas9 mRNA to perform HDR at the CAMK2D locus in mc-iPSCs. sgRNA CAMK-CR1 or CAMK-CR2, Cas9 mRNA and ssODN donor C-CR2 were co-transfected into mc-iPSCs using the EditPro™ RNA transfection reagent (a) The percent wild type and mutant alleles from the transfected cells were detected by droplet digital PCR (ddPCR) using a wild type alleles specific fluorescence probes (VIC) and mutant alleles specific fluorescence probes (FAM), the fluorescence intensity of each droplet in the sample is plotted versus droplet number. Droplets that have fluorescence intensity above the pink threshold line are counted as positive for the target alleles. The bottom panel (green) represents the droplet of wild type alleles while the top panel (blue) represents the alleles that have undergone HDR. The data presented is from one representative experiment using the two sgRNAs at the best concentration of ssOND, 10 pmol. (b) Quantification of mutant allelic frequency. Data presented as mean ± SEM from four independent experiments. sgRNA CAMK-CR2 consistently produced HDR at greater than 15% of the total alleles. (c) Quantification of the intended nucleotide changes by next generation sequencing using the same sgRNA and donor used for the ddPCR experiments. Each bar represents one of the six nucleotide changes included on the donor oligo. The data indicates that complete sequence conversion occurred across the targeted region and at frequencies that were in agreement with the ddPCR results. The two A to G changes not directly measured by ddPCR were also incorporated albeit at lower frequencies compared to those changes that were closer to the CRISPR cleavage site. The data presented is the mean percent intended base alteration at each of their exact genomic coordinates from four independent experiments.
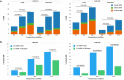
Figure 3
Effects of ‘cold shock’ and ssODN HDR donor designs on HDR efficiencies at the CAMK2D locus in mc-iPSCs as determined by NGS. Various amounts of ssODN C-CR2 or C-CR2-Asym were delivered to mc-iPSCs along with Cas9 mRNA and sgRNA CAMK-CR1 or CAMK-CR2 to achieve HDR at the CAMK2D locus. The experiments were carried out at different temperatures over 24 hour intervals as described in “material and methods”: PL1: 37 °C-37 °C-37 °C, PL2: 37 °C-32 °C-37 °C, PL3: 37 °C-32 °C-32 °C. (a) HDR events using 10 pmol of ssODN HDR donors for each treatment (CAMK-CR1 with C-TR2 or C-TR2-Asym, CAMK-CR2 with C-TR2 or C-TR2-Asym) were determined by NGS as described in “material and methods”. The data presented are the mean percent HDR events (C-CR2: 8 biological replicates from three independent experiments; C-CR2-Asym: 6 biological replicates from two independent experiments). The HDR types were categorized into three groups based on the resulting sequence around the region of the intended mutations. ‘Perfect HDR’: All intended base changes are present with no re-editing indels. Edited HDR: One or more of the intended base changes are present with re-editing indels present. Partial HDR: Some but not all of the intended base changes with no indels. Data demonstrates that increased HDR can be achieved by ‘cold shocking’ the cells and that the majority of the increase is in the ‘Perfect HDR’ category. The significance of total HDR efficiencies difference among the three temperature conditions for each sgRNA and ssODN treatment were analyzed by one way ANOVA (one way ANOVA: P < 0.0001 for all sgRNA and ssODN treatment, P value of follow-up Dunnett’s multiple comparison are shown in the figures). HDR from 30 pmol ssODN and no oligo treatment are shown in the Supplementary Table 3 (b) ‘perfect HDR’ events of each treatment (CAMK-CR1 with C-TR2 or C-TR2-Asym, CAMK-CR2 with C-TR2 or C-TR2-Asym) were plotted to compare ‘perfect HDR’ frequencies between the two ssODN designs. Data presented are the mean percent of ‘perfect HDR’ events ± SEM (6 biological replicates from two independent experiments), The difference of ‘perfect HDR’ frequencies between the two ssODN design in each treatment group were evaluated by Student’s T-test and p values are shown in the figures. (+) strand ssODN C-CR2 promote more ‘perfect HDR’ than (−) strand ssODN C-CR2-Asym across all temperature conditions.
Figure 4
Guide RNAs and single-stranded oligonucleotide (ssODN) donor designs for gene editing at the TGFBR1 locus. (a) Two guide RNAs TR-CR2 and TR-CR3 were designed to specifically target TGFBR1 Exon4 and introduce different sequence alterations by HDR. TR-CR3 is 39 nucleotides downstream of TR-CR2. (b) Two ssODN HDR donors (T-CR2 and T-CR2-Asym) were designed to introduce a silent mutation 12 bp upstream of guide RNA TR-CR2 target site, with three silent mutations within the guide RNA recognition sequence to prevent re-editing of the HDR converted sequence. T-CR2 is a (+) strand HDR donor which is complementary to the guide RNA targeted cleavage strand with balanced homology arms around each side of the intended mutations (5′-73nt and 3′-74nt, respectively). T-CR2-Asym is a (−) strand HDR donor which is complementary to the guide RNA non-targeted strand with homology arms that differ in length (5′-93nt and 3′- 36nt, respectively). (c) Two ssODN donors (T-CR3 and T-CR3-Asym) were designed to introduce a known SNP 12 bp upstream of guide RNA TR-CR3 target site, with two silent mutations within the guide RNA recognition sequence and one silent mutation within the TR-CR3 PAM site to prevent re-editing of the HDR converted sequence. T-CR3 is a (+) strand HDR donor which is complementary to the guide RNA targeted strand with balanced length homology arms (5′-73nt and 3′-72nt, respectively) around each side of the intended mutations. T-CR3-Asym is a (−) strand HDR donor which is complementary to the guide RNA non-targeted strand with unbalanced length homology arms (5′-86nt and 3′-36nt, respectively) around each side of the intended mutations.
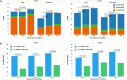
Figure 5
Effects of ‘cold shock’ and ssODN HDR donor designs on HDR efficiency at the TGFBR1 locus in mc-iPSCs. ssODN HDR donors and sgRNAs were co-delivered into mc-iPSCs cells along with Cas9 mRNA to achieve HDR at the TGFBR1 locus. Experiments were carried out at different temperatures over 24 hour intervals as described in “material and methods”: PL1: 37 °C-37 °C-37 °C, PL2: 37 °C-32 °C-37 °C, PL3: 37 °C-32 °C-32 °C. (a) HDR events using 10 pmol of ssODN HDR donors for each treatment (TR-CR2 with T-TR2 or T-TR2-Asym, TR-CR3 with T-TR3 or T-TR3-Asym) were determined by NGS as described in “material and methods”. The data presented are the mean percentage HDR events (4 biological replicates from three independent experiments). The HDR types were categorized into three groups based on the resulting sequence around the region of the intended mutations. ‘Perfect HDR’: All intended base changes are present with no re-editing indels. Edited HDR: One or more of the intended base changes are present with re-editing indels. Partial HDR: Some but not all of the intended base changes with no indels. The significance of total HDR efficiencies differences among the three temperature conditions for each sgRNA and ssODN treatment were analyzed by one way ANOVA (one way ANOVA: TR-CR2 with T-TR2, P = 0.4037; TR-CR2 with T-TR2-Asym, P = 0.0208; TR-CR3 with T-TR3, P = 0.1846; TR-CR3 with T-TR3-Asym, P = 0.0139; P value of follow-up Dunnett’s multiple comparison are shown in the figures). HDR from 30 pmol ssODN and no oligo treatment are shown in the Supplementary Table 4 (b) events using 10 pmol of ssODN HDR donors for each treatment (TR-CR2 with T-TR2 or T-TR2-Asym, TR-CR3 with T-TR3 or T-TR3-Asym) were plotted to compare ‘perfect HDR’ frequencies between the two ssODN designs. Data presented are the mean percent of ‘perfect HDR’ events ± SEM (4 biological replicates from three independent experiments). The difference of ‘perfect HDR’ frequencies between the two ssODN designs in each treatment group were evaluated by Student’s T-test and p values are shown in the figures. Across all temperature conditions, (+) ssODN strand T-CR2 and T-CR3 promote more ‘perfect HDR’ than (−) ssODN strand T-CR2-Asym and T-CR3-Asym, respectively.
Similar articles
- Ectopic expression of RAD52 and dn53BP1 improves homology-directed repair during CRISPR-Cas9 genome editing.
Paulsen BS, Mandal PK, Frock RL, Boyraz B, Yadav R, Upadhyayula S, Gutierrez-Martinez P, Ebina W, Fasth A, Kirchhausen T, Talkowski ME, Agarwal S, Alt FW, Rossi DJ. Paulsen BS, et al. Nat Biomed Eng. 2017 Nov;1(11):878-888. doi: 10.1038/s41551-017-0145-2. Epub 2017 Oct 9. Nat Biomed Eng. 2017. PMID: 31015609 Free PMC article. - Highly efficient genome editing via CRISPR-Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression.
Li XL, Li GH, Fu J, Fu YW, Zhang L, Chen W, Arakaki C, Zhang JP, Wen W, Zhao M, Chen WV, Botimer GD, Baylink D, Aranda L, Choi H, Bechar R, Talbot P, Sun CK, Cheng T, Zhang XB. Li XL, et al. Nucleic Acids Res. 2018 Nov 2;46(19):10195-10215. doi: 10.1093/nar/gky804. Nucleic Acids Res. 2018. PMID: 30239926 Free PMC article. - Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing.
Miyaoka Y, Berman JR, Cooper SB, Mayerl SJ, Chan AH, Zhang B, Karlin-Neumann GA, Conklin BR. Miyaoka Y, et al. Sci Rep. 2016 Mar 31;6:23549. doi: 10.1038/srep23549. Sci Rep. 2016. PMID: 27030102 Free PMC article. - CRISPR/Cas9-mediated correction of human genetic disease.
Men K, Duan X, He Z, Yang Y, Yao S, Wei Y. Men K, et al. Sci China Life Sci. 2017 May;60(5):447-457. doi: 10.1007/s11427-017-9032-4. Epub 2017 May 3. Sci China Life Sci. 2017. PMID: 28534256 Review. - Exploring the potential of genome editing CRISPR-Cas9 technology.
Singh V, Braddick D, Dhar PK. Singh V, et al. Gene. 2017 Jan 30;599:1-18. doi: 10.1016/j.gene.2016.11.008. Epub 2016 Nov 9. Gene. 2017. PMID: 27836667 Review.
Cited by
- GMP-compliant iPS cell lines show widespread plasticity in a new set of differentiation workflows for cell replacement and cancer immunotherapy.
Terheyden-Keighley D, Hühne M, Berger T, Hiller B, Martins S, Gamerschlag A, Sabour D, Meffert A, Kislat A, Slotta C, Hafezi F, Lichte J, Sudheer S, Tessmer K, Psathaki K, Ader M, Kogler G, Greber B. Terheyden-Keighley D, et al. Stem Cells Transl Med. 2024 Sep 10;13(9):898-911. doi: 10.1093/stcltm/szae047. Stem Cells Transl Med. 2024. PMID: 39042522 Free PMC article. - Sharpening the Molecular Scissors: Advances in Gene-Editing Technology.
Broeders M, Herrero-Hernandez P, Ernst MPT, van der Ploeg AT, Pijnappel WWMP. Broeders M, et al. iScience. 2020 Jan 24;23(1):100789. doi: 10.1016/j.isci.2019.100789. Epub 2019 Dec 19. iScience. 2020. PMID: 31901636 Free PMC article. Review. - Microcephaly-associated protein WDR62 shuttles from the Golgi apparatus to the spindle poles in human neural progenitors.
Dell'Amico C, Angulo Salavarria MM, Takeo Y, Saotome I, Dell'Anno MT, Galimberti M, Pellegrino E, Cattaneo E, Louvi A, Onorati M. Dell'Amico C, et al. Elife. 2023 Jun 5;12:e81716. doi: 10.7554/eLife.81716. Elife. 2023. PMID: 37272619 Free PMC article. - Increasing Knockin Efficiency in Mouse Zygotes by Transient Hypothermia.
Bouchareb A, Biggs D, Alghadban S, Preece C, Davies B. Bouchareb A, et al. CRISPR J. 2024 Apr;7(2):111-119. doi: 10.1089/crispr.2023.0077. CRISPR J. 2024. PMID: 38635329 Free PMC article. - Challenges of CRISPR-Based Gene Editing in Primary T Cells.
Rezalotfi A, Fritz L, Förster R, Bošnjak B. Rezalotfi A, et al. Int J Mol Sci. 2022 Feb 1;23(3):1689. doi: 10.3390/ijms23031689. Int J Mol Sci. 2022. PMID: 35163611 Free PMC article. Review.
References
- Bojesen, S. E. et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet45, 371–384, http://www.nature.com/ng/journal/v45/n4/abs/ng.2566.html#supplementary-i... (2013). - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources