Genetic Regulation of the 2D to 3D Growth Transition in the Moss Physcomitrella patens - PubMed (original) (raw)
Genetic Regulation of the 2D to 3D Growth Transition in the Moss Physcomitrella patens
Laura A Moody et al. Curr Biol. 2018.
Abstract
One of the most important events in the history of life on earth was the colonization of land by plants; this transition coincided with and was most likely enabled by the evolution of 3-dimensional (3D) growth. Today, the diverse morphologies exhibited across the terrestrial biosphere arise from the differential regulation of 3D growth processes during development. In many plants, 3D growth is initiated during the first few divisions of the zygote, and therefore, the genetic basis cannot be dissected because mutants do not survive. However, in mosses, which are representatives of the earliest land plants, 3D shoot growth is preceded by a 2D filamentous phase that can be maintained indefinitely. Here, we used the moss Physcomitrella patens to identify genetic regulators of the 2D to 3D transition. Mutant screens yielded individuals that could only grow in 2D, and through an innovative strategy that combined somatic hybridization with bulk segregant analysis and genome sequencing, the causative mutation was identified in one of them. The NO GAMETOPHORES 1 (NOG1) gene, which encodes a ubiquitin-associated protein, is present only in land plant genomes. In mutants that lack PpNOG1 function, transcripts encoding 3D-promoting PpAPB transcription factors [1] are significantly reduced, and apical initial cells specified for 3D growth are not formed. PpNOG1 acts at the earliest identified stage of the 2D to 3D transition, possibly through degradation of proteins that suppress 3D growth. The acquisition of NOG1 function in land plants could thus have enabled the evolution and development of 3D morphology.
Keywords: developmental transitions; land plant evolution; morphogenesis; somatic hybridization.
Copyright © 2018 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
Figure 1
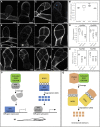
3D Growth Is Abolished in Ppnog1-R Mutants (A–D) 7-day-old (A and C) and 1-month-old (B and D) wild-type (WT) (A and B) and Ppnog1-R (C and D) plants showing protonemal filaments (A and C) and the presence (WT, B) or absence (Ppnog1-R, D) of gametophores. (E) Mean number of gametophores/culture (n = 10) ± SEM (WT = 99.4 ± 5.62; Ppnog1-R = 0 ± 0; t test p < 0.05 ∗∗∗). Scale bars, 100 µm (A and C) and 1 mm (B and D). See Figure S1 for response to cytokinin and auxin.
Figure 2
Identification of the PpNOG1 Locus through Combined Somatic Hybridization and Bulk Segregant Analysis (A–C) Representative phenotype of 1-month-old wild-type Vx plant with gametophores (A), Ppnog1-R plant lacking gametophores (B), and Ppnog1-R/Gd hybrid exhibiting restored gametophore formation (C). Scale bars, 1 mm. (D) Phenotype of progeny derived from three independent Ppnog1-R/Gd hybrid sporophytes. Observed numbers are consistent with the hybrid gametophores being diploid, and the fertilized sporophytes being tetraploid. Chi-square test; p < 0.05 ∗∗∗. (E) Candidate genes in the genetic interval containing the _PpNOG1_ genetic locus. The C > T transition in gene 32970008 (red) generated a premature stop codon. See Figure S2 for overview of strategy.
Figure 3
A Premature Stop Codon in PpNOG1 Abolishes 3D Growth (A) PpNOG1 transcripts in parental Vx and Ppnog1-R. Blocks, exons; asterisks, in-frame stop codon. Scale bar, 1 kb. (B) PpNOG1 protein in Vx contains a ubiquitin-associated domain (UBA), which is missing in Ppnog1-R. # amino acid residues indicated. (C) Mean number of gametophores/culture (n = 10) in Vx, nog1-R, and five independent Ppnog1-R lines complemented with full-length PpNOG1 cDNA. Error bars ± SEM. (D–G) 1-month-old Vx (D), Ppnog1-R (E), complemented Ppnog1-R (F), Ppnog1-D1 (G), and Ppnog1-D2 (H) disruptant mutants. Scale bars, 1 mm. See Figure S3 and Data S1 for details of complementation and disruptant constructs.
Figure 4
PpNOG1 Function (A–L) Propidium iodide stained Vx (A–D) and Ppnog1-R (E–L) side-branch cells (A and E) and buds at 2- (B and F), 3- (C, G, and H), 4- (D, I, and J), and 5- (K and L) cell stages. Scale bars, 10 μm. •, gametophore initial. (M–O) Relative transcript levels in protonemata: PpNOG1 in wild-type ± BAP and/or NAA (M) and PpAPB1-4 in Vx and the Ppnog1-D1 and the Ppnog1-D2 disruptants (N and O). ANOVA, ∗∗∗p < 0.05. (P and Q) Model for PpNOG1 function: PpDEK1 and PpNOG1 act antagonistically during side-branch initiation to regulate PpAPB transcription (P) and then act together to enable divisions that produce the gametophore initial (Q). See Figure S4 for NOG1 phylogeny.
Similar articles
- The 2D to 3D growth transition in the moss Physcomitrella patens.
Moody LA. Moody LA. Curr Opin Plant Biol. 2019 Feb;47:88-95. doi: 10.1016/j.pbi.2018.10.001. Epub 2018 Nov 3. Curr Opin Plant Biol. 2019. PMID: 30399606 Review. - NO GAMETOPHORES 2 Is a Novel Regulator of the 2D to 3D Growth Transition in the Moss Physcomitrella patens.
Moody LA, Kelly S, Clayton R, Weeks Z, Emms DM, Langdale JA. Moody LA, et al. Curr Biol. 2021 Feb 8;31(3):555-563.e4. doi: 10.1016/j.cub.2020.10.077. Epub 2020 Nov 25. Curr Biol. 2021. PMID: 33242390 - CLAVATA Was a Genetic Novelty for the Morphological Innovation of 3D Growth in Land Plants.
Whitewoods CD, Cammarata J, Nemec Venza Z, Sang S, Crook AD, Aoyama T, Wang XY, Waller M, Kamisugi Y, Cuming AC, Szövényi P, Nimchuk ZL, Roeder AHK, Scanlon MJ, Harrison CJ. Whitewoods CD, et al. Curr Biol. 2018 Aug 6;28(15):2365-2376.e5. doi: 10.1016/j.cub.2018.05.068. Epub 2018 Jul 19. Curr Biol. 2018. PMID: 30033333 Free PMC article. - PpGRAS12 acts as a positive regulator of meristem formation in Physcomitrium patens.
Beheshti H, Strotbek C, Arif MA, Klingl A, Top O, Frank W. Beheshti H, et al. Plant Mol Biol. 2021 Nov;107(4-5):293-305. doi: 10.1007/s11103-021-01125-z. Epub 2021 Feb 17. Plant Mol Biol. 2021. PMID: 33598827 Free PMC article. - Eight types of stem cells in the life cycle of the moss Physcomitrella patens.
Kofuji R, Hasebe M. Kofuji R, et al. Curr Opin Plant Biol. 2014 Feb;17:13-21. doi: 10.1016/j.pbi.2013.10.007. Epub 2013 Nov 16. Curr Opin Plant Biol. 2014. PMID: 24507489 Review.
Cited by
- Physcomitrium patens: A Single Model to Study Oriented Cell Divisions in 1D to 3D Patterning.
de Keijzer J, Freire Rios A, Willemsen V. de Keijzer J, et al. Int J Mol Sci. 2021 Mar 5;22(5):2626. doi: 10.3390/ijms22052626. Int J Mol Sci. 2021. PMID: 33807788 Free PMC article. Review. - Spindle motility skews division site determination during asymmetric cell division in Physcomitrella.
Kozgunova E, Yoshida MW, Reski R, Goshima G. Kozgunova E, et al. Nat Commun. 2022 May 5;13(1):2488. doi: 10.1038/s41467-022-30239-1. Nat Commun. 2022. PMID: 35513464 Free PMC article. - The Moss Physcomitrium (Physcomitrella) patens: A Model Organism for Non-Seed Plants.
Rensing SA, Goffinet B, Meyberg R, Wu SZ, Bezanilla M. Rensing SA, et al. Plant Cell. 2020 May;32(5):1361-1376. doi: 10.1105/tpc.19.00828. Epub 2020 Mar 9. Plant Cell. 2020. PMID: 32152187 Free PMC article. Review. - Unravelling 3D growth in the moss Physcomitrium patens.
Moody LA. Moody LA. Essays Biochem. 2022 Dec 8;66(6):769-779. doi: 10.1042/EBC20220048. Essays Biochem. 2022. PMID: 36342774 Free PMC article. - The hornworts: morphology, evolution and development.
Frangedakis E, Shimamura M, Villarreal JC, Li FW, Tomaselli M, Waller M, Sakakibara K, Renzaglia KS, Szövényi P. Frangedakis E, et al. New Phytol. 2021 Jan;229(2):735-754. doi: 10.1111/nph.16874. Epub 2020 Sep 15. New Phytol. 2021. PMID: 32790880 Free PMC article. Review.
References
- Aoyama T., Hiwatashi Y., Shigyo M., Kofuji R., Kubo M., Ito M., Hasebe M. AP2-type transcription factors determine stem cell identity in the moss Physcomitrella patens. Development. 2012;139:3120–3129. - PubMed
- Cove D., Bezanilla M., Harries P., Quatrano R. Mosses as model systems for the study of metabolism and development. Annu. Rev. Plant Biol. 2006;57:497–520. - PubMed
- Harrison C.J., Roeder A.H., Meyerowitz E.M., Langdale J.A. Local cues and asymmetric cell divisions underpin body plan transitions in the moss Physcomitrella patens. Curr. Biol. 2009;19:461–471. - PubMed
- Kofuji R., Hasebe M. Eight types of stem cells in the life cycle of the moss Physcomitrella patens. Curr. Opin. Plant Biol. 2014;17:13–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources