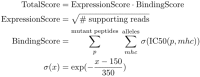Computational Pipeline for the PGV-001 Neoantigen Vaccine Trial - PubMed (original) (raw)
Computational Pipeline for the PGV-001 Neoantigen Vaccine Trial
Alex Rubinsteyn et al. Front Immunol. 2018.
Abstract
This paper describes the sequencing protocol and computational pipeline for the PGV-001 personalized vaccine trial. PGV-001 is a therapeutic peptide vaccine targeting neoantigens identified from patient tumor samples. Peptides are selected by a computational pipeline that identifies mutations from tumor/normal exome sequencing and ranks mutant sequences by a combination of predicted Class I MHC affinity and abundance estimated from tumor RNA. The personalized genomic vaccine (PGV) pipeline is modular and consists of independently usable tools and software libraries. We hope that the functionality of these tools may extend beyond the specifics of the PGV-001 trial and enable other research groups in their own neoantigen investigations.
Keywords: computational pipeline; genomics; immunoinformatics; neoantigens; personalized vaccine.
Figures
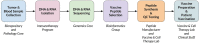
Figure 1
Overview of PGV-001 trial.
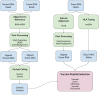
Figure 2
Schematic of bioinformatics tools used in PGV-001 pipeline.
Figure 3
Overview of Isovar algorithm for determining mutant protein sequences.
Figure 4
Schematic representation of a somatic mutation co-occurring with a germline mutation.
Figure 5
Screenshot from IGV with tumor DNA on top and tumor RNA on bottom. The two somatic variants from patient data 7 amino acids apart. If these mutations were considered without phasing, we would get two different vaccine peptides, neither of which would match the protein sequence produced by tumor cells.
Figure 6
TotalScore used to rank somatic variants in a way that attempts to balance predict MHC binding and abundance. ExpressionScore uses read count (instead of a normalized measure like FPKM) since these scoring criteria are not meant to be compared between patient samples. BindingScore sums normalized binding affinities of mutant peptides across all patient alleles and lengths between 8 and 11.
Similar articles
- OpenVax: An Open-Source Computational Pipeline for Cancer Neoantigen Prediction.
Kodysh J, Rubinsteyn A. Kodysh J, et al. Methods Mol Biol. 2020;2120:147-160. doi: 10.1007/978-1-0716-0327-7_10. Methods Mol Biol. 2020. PMID: 32124317 - Identification of Individual Cancer-Specific Somatic Mutations for Neoantigen-Based Immunotherapy of Lung Cancer.
Karasaki T, Nagayama K, Kawashima M, Hiyama N, Murayama T, Kuwano H, Nitadori J, Anraku M, Sato M, Miyai M, Hosoi A, Matsushita H, Kikugawa S, Matoba R, Ohara O, Kakimi K, Nakajima J. Karasaki T, et al. J Thorac Oncol. 2016 Mar;11(3):324-33. doi: 10.1016/j.jtho.2015.11.006. Epub 2015 Dec 29. J Thorac Oncol. 2016. PMID: 26752676 - Detection of neoantigen-specific T cells following a personalized vaccine in a patient with glioblastoma.
Johanns TM, Miller CA, Liu CJ, Perrin RJ, Bender D, Kobayashi DK, Campian JL, Chicoine MR, Dacey RG, Huang J, Fritsch EF, Gillanders WE, Artyomov MN, Mardis ER, Schreiber RD, Dunn GP. Johanns TM, et al. Oncoimmunology. 2019 Jan 25;8(4):e1561106. doi: 10.1080/2162402X.2018.1561106. eCollection 2019. Oncoimmunology. 2019. PMID: 30906654 Free PMC article. - [Neoantigens and Whole-Exome Sequencing].
Karasaki T, Nakajima J, Kakimi K. Karasaki T, et al. Gan To Kagaku Ryoho. 2016 Jul;43(7):791-7. Gan To Kagaku Ryoho. 2016. PMID: 27431622 Review. Japanese. - Main Strategies for the Identification of Neoantigens.
Gopanenko AV, Kosobokova EN, Kosorukov VS. Gopanenko AV, et al. Cancers (Basel). 2020 Oct 7;12(10):2879. doi: 10.3390/cancers12102879. Cancers (Basel). 2020. PMID: 33036391 Free PMC article. Review.
Cited by
- Molecular targets and strategies in the development of nucleic acid cancer vaccines: from shared to personalized antigens.
Chi WY, Hu Y, Huang HC, Kuo HH, Lin SH, Kuo CJ, Tao J, Fan D, Huang YM, Wu AA, Hung CF, Wu TC. Chi WY, et al. J Biomed Sci. 2024 Oct 9;31(1):94. doi: 10.1186/s12929-024-01082-x. J Biomed Sci. 2024. PMID: 39379923 Free PMC article. Review. - Developing Vaccines in Pancreatic Adenocarcinoma: Trials and Tribulations.
Phan T, Fan D, Melstrom LG. Phan T, et al. Curr Oncol. 2024 Aug 23;31(9):4855-4884. doi: 10.3390/curroncol31090361. Curr Oncol. 2024. PMID: 39329989 Free PMC article. Review. - Current Trends and Innovative Approaches in Cancer Immunotherapy.
Kim J, Maharjan R, Park J. Kim J, et al. AAPS PharmSciTech. 2024 Jul 24;25(6):168. doi: 10.1208/s12249-024-02883-x. AAPS PharmSciTech. 2024. PMID: 39044047 Review. - Transformers meets neoantigen detection: a systematic literature review.
Machaca V, Goyzueta V, Cruz MG, Sejje E, Pilco LM, López J, Túpac Y. Machaca V, et al. J Integr Bioinform. 2024 Jul 4;21(2):20230043. doi: 10.1515/jib-2023-0043. eCollection 2024 Jun 1. J Integr Bioinform. 2024. PMID: 38960869 Free PMC article. - Structural basis for self-discrimination by neoantigen-specific TCRs.
Finnigan JP, Newman JH, Patskovsky Y, Patskovska L, Ishizuka AS, Lynn GM, Seder RA, Krogsgaard M, Bhardwaj N. Finnigan JP, et al. Nat Commun. 2024 Mar 8;15(1):2140. doi: 10.1038/s41467-024-46367-9. Nat Commun. 2024. PMID: 38459027 Free PMC article.
References
- Finnigan JP, Jr, Rubinsteyn A, Hammerbacher J, Bhardwaj N. Mutation-derived tumor antigens: novel targets in cancer immunotherapy. Oncology (2015) 29:970–2, 974–5. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials