Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis - PubMed (original) (raw)
Review
Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis
Mikael S Lindström et al. Oncogene. 2018 May.
Abstract
The nucleolus is the major site for synthesis of ribosomes, complex molecular machines that are responsible for protein synthesis. A wealth of research over the past 20 years has clearly indicated that both quantitative and qualitative alterations in ribosome biogenesis can drive the malignant phenotype via dysregulation of protein synthesis. However, numerous recent proteomic, genomic, and functional studies have implicated the nucleolus in the regulation of processes that are unrelated to ribosome biogenesis, including DNA-damage response, maintenance of genome stability and its spatial organization, epigenetic regulation, cell-cycle control, stress responses, senescence, global gene expression, as well as assembly or maturation of various ribonucleoprotein particles. In this review, the focus will be on features of rDNA genes, which make them highly vulnerable to DNA damage and intra- and interchromosomal recombination as well as built-in mechanisms that prevent and repair rDNA damage, and how dysregulation of this interplay affects genome-wide DNA stability, gene expression and the balance between euchromatin and heterochromatin. We will also present the most recent insights into how malfunction of these cellular processes may be a central driving force of human malignancies, and propose a promising new therapeutic approach for the treatment of cancer.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
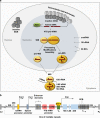
Fig. 1
Nucleolus and rDNA genes. a Intranucleolar heterochromatic inactive rDNA repeats are tightly connected with perinucleolar heterochromatin, whereas euchromatic active rDNA genes are transcribed into the 47 S pre-rRNA by Pol I. The 47 S pre-rRNA is co-transcriptionally assembled into the 90 S processome with RPs and 5 S rRNA and modified by ~200 snoRNPs. Following cleavage of the pre-rRNA, the 90 S processome separates into a pre-60S subunit and a pre-40S subunit that largely follow independent maturation pathways in the nucleolus, the nucleoplasm and the cytoplasm. Modifications of rRNA are indicated with lollipops. b Schematic diagram of the rDNA array. Shown are coding regions for 18 S, 5.8 S, and 28 S rRNA as well as IGS with the rDNA core promoter and upstream control element (UCE), enhancers, spacer promoter, and Pol I promoter, ORIs as well as T0 that binds TTF-I and T1, T4, and T5 that act as RFBs. DJ and PJ sequences that flank the rDNA array are indicated
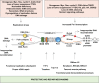
Fig. 2
Causes and consequences of conflicts between Pol I transcriptional and replication machineries. The indicated oncogenic signaling pathways can lead an aberrant increase in the rate of Pol I transcription. Convergence of Pol I and replication machineries in head-on orientation leads to the accumulation of positive DNA supercoiling, which slows down both machineries. Negative DNA supercoiling forming behind transcription bubble leads to opening of rDNA strands and the formation of highly stable rRNA:rDNA hybrids, leaving the displaced non-template strand. The rDNA genes are predilection sites for the formation of R- loops owing to the presence of a region of GC skewing, downstream of unmethylated CpG island rDNA promoters. Clashes of Pol I transcriptional machinery and R-loops with replication fork may cause its stalling or collapse. Perturbing replication of rDNA may further potentiate conflicts between replicating forks and Pol I transcriptional machinery as well as generation and stabilization of co-transcriptional R-loops and R-loop dependent rDNA damage. Several protective and DNA repair mechanisms are shown that preserve genome stability upon conflicts between Pol I transcriptional and replication machineries, thus preventing tumorigenesis. Asterisks indicate DNA damage
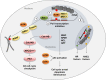
Fig. 3
Nucleolar response to nuclear or nucleolar DNA damage. DSBs in the nuclear chromatin cause Pol I inhibition in the nucleolus through an ATM-dependent signaling pathway, which includes NBS1, PARP, and Treacle. Pol I is also inhibited upon direct rDNA damage in an ATM-NBS1-MDC1-dependent manner. Inhibition of Pol I transcription is most likely responsible for the formation of nucleolar caps and relocalization of damaged rDNA to this structures, which are enriched in HR and NHEJ factors, thus providing an optimal environment for proper rDNA repair. Notably, cap formation was not observed upon nuclear DNA damage is several studies, in spite of Pol I inhibition. DDR and associated Pol I inhibition activate p53 through the impaired ribosome biogenesis checkpoint (IRBC), NMP1, NCL, ARF, ATM, and ATR. Upon genotoxic stress, Cdc14B is released from the nucleolus to the nucleoplasm where it promotes APC/CCdh1-dependent Plk1 degradation, triggering a G2-phase cell-cycle checkpoint. Asterisks indicate DSBs
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.
Ryan R, Hill S. Ryan R, et al. Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article. - Australia in 2030: what is our path to health for all?
Backholer K, Baum F, Finlay SM, Friel S, Giles-Corti B, Jones A, Patrick R, Shill J, Townsend B, Armstrong F, Baker P, Bowen K, Browne J, Büsst C, Butt A, Canuto K, Canuto K, Capon A, Corben K, Daube M, Goldfeld S, Grenfell R, Gunn L, Harris P, Horton K, Keane L, Lacy-Nichols J, Lo SN, Lovett RW, Lowe M, Martin JE, Neal N, Peeters A, Pettman T, Thoms A, Thow AMT, Timperio A, Williams C, Wright A, Zapata-Diomedi B, Demaio S. Backholer K, et al. Med J Aust. 2021 May;214 Suppl 8:S5-S40. doi: 10.5694/mja2.51020. Med J Aust. 2021. PMID: 33934362 - "It Is a Big Spider Web of Things": Sensory Experiences of Autistic Adults in Public Spaces.
MacLennan K, Woolley C, Andsensory E, Heasman B, Starns J, George B, Manning C. MacLennan K, et al. Autism Adulthood. 2023 Dec 1;5(4):411-422. doi: 10.1089/aut.2022.0024. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116051 Free PMC article. - Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.
Triana L, Palacios Huatuco RM, Campilgio G, Liscano E. Triana L, et al. Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
- STAU2 protein level is controlled by caspases and the CHK1 pathway and regulates cell cycle progression in the non-transformed hTERT-RPE1 cells.
Condé L, Gonzalez Quesada Y, Bonnet-Magnaval F, Beaujois R, DesGroseillers L. Condé L, et al. BMC Mol Cell Biol. 2021 Mar 4;22(1):16. doi: 10.1186/s12860-021-00352-y. BMC Mol Cell Biol. 2021. PMID: 33663378 Free PMC article. - The Nucleolus and Its Interactions with Viral Proteins Required for Successful Infection.
Ulloa-Aguilar JM, Herrera Moro Huitron L, Benítez-Zeferino RY, Cerna-Cortes JF, García-Cordero J, León-Reyes G, Guzman-Bautista ER, Farfan-Morales CN, Reyes-Ruiz JM, Miranda-Labra RU, De Jesús-González LA, León-Juárez M. Ulloa-Aguilar JM, et al. Cells. 2024 Sep 21;13(18):1591. doi: 10.3390/cells13181591. Cells. 2024. PMID: 39329772 Free PMC article. Review. - Translational Control during Cellular Senescence.
Payea MJ, Anerillas C, Tharakan R, Gorospe M. Payea MJ, et al. Mol Cell Biol. 2021 Jan 25;41(2):e00512-20. doi: 10.1128/MCB.00512-20. Print 2021 Jan 25. Mol Cell Biol. 2021. PMID: 33077499 Free PMC article. Review. - Nuclear Organization in Response to Stress: A Special Focus on Nucleoli.
Batnasan E, Koivukoski S, Kärkkäinen M, Latonen L. Batnasan E, et al. Results Probl Cell Differ. 2022;70:469-494. doi: 10.1007/978-3-031-06573-6_17. Results Probl Cell Differ. 2022. PMID: 36348119 - Loss of Peter Pan (PPAN) Affects Mitochondrial Homeostasis and Autophagic Flux.
Dannheisig DP, Beck E, Calzia E, Walther P, Behrends C, Pfister AS. Dannheisig DP, et al. Cells. 2019 Aug 14;8(8):894. doi: 10.3390/cells8080894. Cells. 2019. PMID: 31416196 Free PMC article.
References
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, et al. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous