Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation - PubMed (original) (raw)
Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation
Tao Li et al. Nucleic Acids Res. 2018.
Abstract
UHRF1 plays multiple roles in regulating DNMT1-mediated DNA methylation maintenance during DNA replication. The UHRF1 C-terminal RING finger functions as an ubiquitin E3 ligase to establish histone H3 ubiquitination at Lys18 and/or Lys23, which is subsequently recognized by DNMT1 to promote its localization onto replication foci. Here, we present the crystal structure of DNMT1 RFTS domain in complex with ubiquitin and highlight a unique ubiquitin binding mode for the RFTS domain. We provide evidence that UHRF1 N-terminal ubiquitin-like domain (UBL) also binds directly to DNMT1. Despite sharing a high degree of structural similarity, UHRF1 UBL and ubiquitin bind to DNMT1 in a very distinct fashion and exert different impacts on DNMT1 enzymatic activity. We further show that the UHRF1 UBL-mediated interaction between UHRF1 and DNMT1, and the binding of DNMT1 to ubiquitinated histone H3 that is catalyzed by UHRF1 RING domain are critical for the proper subnuclear localization of DNMT1 and maintenance of DNA methylation. Collectively, our study adds another layer of complexity to the regulatory mechanism of DNMT1 activation by UHRF1 and supports that individual domains of UHRF1 participate and act in concert to maintain DNA methylation patterns.
Figures
Figure 1.
Molecular determinants of DNMT1 binding to ubiquitin and UBL domain of UHRF1. (A) Domain organization of DNMT1. Three DNMT1 fragments were prepared for ubiquitin binding analysis. (B and C) 15N HSQC spectra show that ubiquitin binds to DNMT1 RFTS domain (aa: 351–600) but not DNMT1_621 (aa: 621–1616), as illustrated by strong selective line broadening effect induced by addition of DNMT1 RFTS domain but not DNMT1_621. (D) Domain organization of DNMT1 fragments that were prepared for UHRF1 UBL binding. (E and F) NMR titration indicates UHRF1 UBL binds to DNMT1 RFTS domain as well as DNMT1_621, as illustrated by strong line broadening of certain resonances induced by the binding of DNMT1 RFTS (E) and DNMT1_621 fragment (F). (G and H) ITC confirms the molecular interaction of DNMT1 RFTS domain with ubiquitin and UHRF1 UBL domain. The top panel shows experimental ITC curve of titrating DNMT1 RFTS domain into ubiquitin (G) and UHRF1 UBL (H) respectively. The lower panel shows fitted curves of calorimetric titrations. The measured dissociation constant (_K_d) is shown.
Figure 2.
Crystal structure of DNMT1 RFTS domain in complex with ubiquitin. (A) 1:2 DNMT1 RFTS domain-ubiquitin contents of the asymmetric unit are shown as ribbon diagrams. DNMT1 RFTS domain is colored in green, two ubiquitin molecules are colored in marine and cyan respectively. (B and C) Close-up view of the interactions between the RFTS domain and ubiquitin 1 (Ubq1). (D and E) Detailed view of the interaction network of the RFTS domain with ubiquitin 2 (Ubq2). Residues that form the binding interface are depicted as stick models and labeled. Hydrogen bonds and salt bridges are shown in magenta dashes. (F) DNMT1 RFTS domain undergoes conformational changes upon ubiquitin binding. The α-helix (aa: 493–518) of RFTS domain adopts as a straight α-helical conformation in DNMT1 (PDB ID: 4WXX) but bends about 32° in the middle of α-helix (aa: 493–518) at the position of Met502 in RFTS/ubiquitin complex. Ubiquitin binding caused the bending of RFTS α-helix (aa: 493–518) that connects the preceding β-barrel (aa: 400–490) to the α-helical bundle, this eventually leads to large changes in the relative orientation of the β-barrel with respect to α-helical bundle.
Figure 3.
Experimental measurement of DNMT1 RFTS domain binding to ubiquitin or UHRF1 UBL domain. (A) ITC fitting curves for binding of DNMT1 RFTS domain and its mutants to ubiquitin, the insert lists the calculated dissociation constant (_K_d). ND, not detectable. (B) ITC fitting curves for binding of DNMT1 RFTS domain to ubiquitin wild-type and mutants, along with the calculated _K_d. (C) ITC fitting curves for binding of DNMT1 RFTS domain and its mutants with UHRF1 UBL domain with dissociation constant (_K_d) values indicated. (D and E) In vitro DNA methylation reactions were performed as a function of time and the concentration of ubiquitin or UHRF1 UBL. (D) DNMT1_351 (aa: 351–1616) or (E) DNMT1_621 (aa: 621–1616) was used as the enzyme, and a 30-bp long hemimethylated DNA fragment as the substrate. (F and G) In vitro DNA methylation reactions were performed as a function of time and the concentration of ubiquitin, histone H3 or H3ub2. (F) DNMT1_351 (aa: 351–1616) or (G) DNMT1_621 (aa: 621–1616) was used as the enzyme.
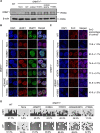
Figure 4.
Assessing nuclear localization and DNA methylation status in DNMT1−/− mouse embryonic stem cells stably expressing DNMT1 wild-type or mutants. (A) Flag-Myc-tagged wild-type DNMT1, DNMT1 E384A, E397A, E384A/E397A and Y399A mutants were stably expressed in DNMT1−/− mouse ESCs, as analyzed by western blot using antibodies that recognize DNMT1. The expression level of these exogenous proteins similar to endogenous DNMT1 was selected for the subsequent study. β-actin was selected as a loading control. (B) Immunofluorescence analysis of DNMT1 focal staining pattern in DNMT1 wild-type or knockout mouse ES cells or DNMT1−/− ES cells stably expressing DNMT1 wild-type, E384A or E397A or Y339G single point mutant, or E384A/E397A double point mutant respectively. Scale bars, 10 μm. (C) Immunostaining using an antibody against 5mC in control and DNMT1−/− mouse ES cells after genetic complementation with DNMT1 wild-type or various mutants. 5mC fluorescence signals from ∼50 cells were quantified and normalized against the wild-type cells, the mean value with a standard error has been provided. (D) The DNA methylation status of LINE1 and IAP was analyzed by bisulfite sequencing in control, DNMT1−/− ESCs and DNMT1−/− ESCs stably expressing DNMT1 wild-type, E384A, E397A, E384A/E397A and Y399A mutants. The percentage of 5mC was calculated and shown.
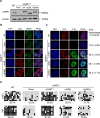
Figure 5.
Both UHRF1 UBL and RING finger are critical for DNMT1 proper nuclear localization and maintenance DNA methylation in mouse embryonic stem cells. (A) UHRF1−/− mouse ES cell lines stably expressing Flag-Myc-tagged UHRF1 wild-type or various mutants were analyzed by western blot using antibodies that recognize UHRF1. Flag-Myc-tagged UHRF1 or mutants expressed at a level similar to endogenous UHRF1 were selected for the study. β-actin was selected as a loading control. (B) Immunofluorescence analysis of DNMT1 focal staining pattern in UHRF1 wild-type, UHRF1 knockout mouse ES cells and UHRF1−/− ES cells transfected with UHRF1-ΔUBL or UHRF1-ΔRING truncated mutant. Exogenous expression of UHRF1 and mutants was detected by Flag antibodies. Scale bars, 10 μm. (C) Immunostaining using an antibody against 5mC in control and UHRF1−/− mouse ES cells after genetic complementation with wild-type or UHRF1-ΔUBL or UHRF1-ΔRING. The 5mC levels relative to wild-type ESCs were shown. Error bars represent ± s.e.m. (D) The DNA methylation status of LINE1 and IAP was analyzed by bisulfite sequencing in wild-type ESCs (as control), UHRF1−/− ESCs and UHRF1−/- ESCs stably expressing UHRF1 wild-type, or UHRF1-ΔUBL or UHRF1-ΔRING mutants. The percentage of 5mC was calculated and shown.
Similar articles
- Enhanced processivity of Dnmt1 by monoubiquitinated histone H3.
Mishima Y, Brueckner L, Takahashi S, Kawakami T, Otani J, Shinohara A, Takeshita K, Garvilles RG, Watanabe M, Sakai N, Takeshima H, Nachtegael C, Nishiyama A, Nakanishi M, Arita K, Nakashima K, Hojo H, Suetake I. Mishima Y, et al. Genes Cells. 2020 Jan;25(1):22-32. doi: 10.1111/gtc.12732. Epub 2019 Dec 3. Genes Cells. 2020. PMID: 31680384 - Critical Role of the UBL Domain in Stimulating the E3 Ubiquitin Ligase Activity of UHRF1 toward Chromatin.
Foster BM, Stolz P, Mulholland CB, Montoya A, Kramer H, Bultmann S, Bartke T. Foster BM, et al. Mol Cell. 2018 Nov 15;72(4):739-752.e9. doi: 10.1016/j.molcel.2018.09.028. Epub 2018 Nov 1. Mol Cell. 2018. PMID: 30392929 Free PMC article. - DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination.
Qin W, Wolf P, Liu N, Link S, Smets M, La Mastra F, Forné I, Pichler G, Hörl D, Fellinger K, Spada F, Bonapace IM, Imhof A, Harz H, Leonhardt H. Qin W, et al. Cell Res. 2015 Aug;25(8):911-29. doi: 10.1038/cr.2015.72. Epub 2015 Jun 12. Cell Res. 2015. PMID: 26065575 Free PMC article. - Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns.
Bronner C, Alhosin M, Hamiche A, Mousli M. Bronner C, et al. Genes (Basel). 2019 Jan 18;10(1):65. doi: 10.3390/genes10010065. Genes (Basel). 2019. PMID: 30669400 Free PMC article. Review. - Regulatory mechanism and biological function of UHRF1-DNMT1-mediated DNA methylation.
Ren Y. Ren Y. Funct Integr Genomics. 2022 Dec;22(6):1113-1126. doi: 10.1007/s10142-022-00918-9. Epub 2022 Nov 14. Funct Integr Genomics. 2022. PMID: 36372834 Review.
Cited by
- Mechanism of nucleosomal H2A K13/15 monoubiquitination and adjacent dual monoubiquitination by RNF168.
Ai H, Tong Z, Deng Z, Shi Q, Tao S, Sun G, Liang J, Sun M, Wu X, Zheng Q, Liang L, Yin H, Li JB, Gao S, Tian C, Liu L, Pan M. Ai H, et al. Nat Chem Biol. 2024 Oct 11. doi: 10.1038/s41589-024-01750-x. Online ahead of print. Nat Chem Biol. 2024. PMID: 39394267 - STUB1-mediated K63-linked ubiquitination of UHRF1 promotes the progression of cholangiocarcinoma by maintaining DNA hypermethylation of PLA2G2A.
Chen J, Wang D, Wu G, Xiong F, Liu W, Wang Q, Kuai Y, Huang W, Qi Y, Wang B, Chen Y. Chen J, et al. J Exp Clin Cancer Res. 2024 Sep 13;43(1):260. doi: 10.1186/s13046-024-03186-6. J Exp Clin Cancer Res. 2024. PMID: 39267107 Free PMC article. - Structural insight into the DNMT1 reaction cycle by cryo-electron microscopy.
De I, Weidenhausen J, Concha N, Müller CW. De I, et al. PLoS One. 2024 Sep 3;19(9):e0307850. doi: 10.1371/journal.pone.0307850. eCollection 2024. PLoS One. 2024. PMID: 39226277 Free PMC article. - Oncogenic Roles of UHRF1 in Cancer.
Kim A, Benavente CA. Kim A, et al. Epigenomes. 2024 Jul 1;8(3):26. doi: 10.3390/epigenomes8030026. Epigenomes. 2024. PMID: 39051184 Free PMC article. Review. - Mechanistic insights into UHRF1‑mediated DNA methylation by structure‑based functional clarification of UHRF1 domains (Review).
Song Y, Liu H, Xian Q, Gui C, Xu M, Zhou Y. Song Y, et al. Oncol Lett. 2023 Nov 2;26(6):542. doi: 10.3892/ol.2023.14129. eCollection 2023 Dec. Oncol Lett. 2023. PMID: 38020304 Free PMC article. Review.
References
- Smith Z.D., Meissner A.. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013; 14:204–220. - PubMed
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007; 8:286–298. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources