Electron Transfer to Nitrogenase in Different Genomic and Metabolic Backgrounds - PubMed (original) (raw)
. 2018 Apr 24;200(10):e00757-17.
doi: 10.1128/JB.00757-17. Print 2018 May 15.
Affiliations
- PMID: 29483165
- PMCID: PMC5915786
- DOI: 10.1128/JB.00757-17
Electron Transfer to Nitrogenase in Different Genomic and Metabolic Backgrounds
Saroj Poudel et al. J Bacteriol. 2018.
Abstract
Nitrogenase catalyzes the reduction of dinitrogen (N2) using low-potential electrons from ferredoxin (Fd) or flavodoxin (Fld) through an ATP-dependent process. Since its emergence in an anaerobic chemoautotroph, this oxygen (O2)-sensitive enzyme complex has evolved to operate in a variety of genomic and metabolic backgrounds, including those of aerobes, anaerobes, chemotrophs, and phototrophs. However, whether pathways of electron delivery to nitrogenase are influenced by these different metabolic backgrounds is not well understood. Here, we report the distribution of homologs of Fds, Flds, and Fd-/Fld-reducing enzymes in 359 genomes of putative N2 fixers (diazotrophs). Six distinct lineages of nitrogenase were identified, and their distributions largely corresponded to differences in the host cells' ability to integrate O2 or light into energy metabolism. The predicted pathways of electron transfer to nitrogenase in aerobes, facultative anaerobes, and phototrophs varied from those in anaerobes at the levels of Fds/Flds used to reduce nitrogenase, the enzymes that generate reduced Fds/Flds, and the putative substrates of these enzymes. Proteins that putatively reduce Fd with hydrogen or pyruvate were enriched in anaerobes, while those that reduce Fd with NADH/NADPH were enriched in aerobes, facultative anaerobes, and anoxygenic phototrophs. The energy metabolism of aerobic, facultatively anaerobic, and anoxygenic phototrophic diazotrophs often yields reduced NADH/NADPH that is not sufficiently reduced to drive N2 reduction. At least two mechanisms have been acquired by these taxa to overcome this limitation and to generate electrons with potentials capable of reducing Fd. These include the bifurcation of electrons or the coupling of Fd reduction to reverse ion translocation.IMPORTANCE Nitrogen fixation supplies fixed nitrogen to cells from a variety of genomic and metabolic backgrounds, including those of aerobes, facultative anaerobes, chemotrophs, and phototrophs. Here, using informatics approaches applied to genomic data, we show that pathways of electron transfer to nitrogenase in metabolically diverse diazotrophic taxa have diversified primarily in response to host cells' acquired ability to integrate O2 or light into their energy metabolism. The acquisition of two key enzyme complexes enabled aerobic and facultatively anaerobic phototrophic taxa to generate electrons of sufficiently low potential to reduce nitrogenase: the bifurcation of electrons via the Fix complex or the coupling of Fd reduction to reverse ion translocation via the Rhodobacter nitrogen fixation (Rnf) complex.
Keywords: Rnf; bifurcation; ferredoxin; fix; flavodoxin; hydrogen; nitrogen fixation; nitrogenase; oxygen; photosynthesis; pyruvate.
Copyright © 2018 American Society for Microbiology.
Figures
FIG 1
Phylogenetic reconstruction of a concatenation of H, D, and K subunits of nitrogenase and uncharacterized nitrogenase paralogs (n = 420 concatenated protein sequences). All nodes shown exhibited bootstrap supports of >90% (out of 1,000 bootstrap replicates), except where a black box (>70%) is shown. Nif, molybdenum (Mo) nitrogenase; Anf, iron-only (Fe) nitrogenase; Vnf, vanadium (V) nitrogenase; Unc, uncharacterized nitrogenase-like proteins.
FIG 2
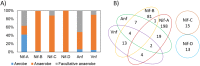
Homologs of nitrogenase identified in the genomes of putative diazotrophs. (A) Histogram depicting the percentage of diazotrophs within each specified Nif lineage (see Fig. 1) that are aerobic, anaerobic, or facultatively anaerobic, as determined from surveys of the literature for cultivated organisms. (B) Venn diagram representing the numbers of genomes that encode one or more specified lineages of nitrogenase (see Fig. 1). Genomes that encode Nif-C and Nif-D do not encode other isoforms of nitrogenase and are thus depicted as separate.
FIG 3
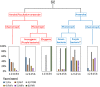
Flow chart of nitrogenase in different metabolic backgrounds and histograms depicting the percentage of enzyme homologs putatively involved in Fd/Fld reduction identified in the genomes comprising a specified group. Aerobic/facultative anaerobic and anaerobic organisms are further classified as phototrophs or chemotrophs. Phototrophs in aerobes/facultative anaerobes are further classified based on whether they are oxygenic or anoxygenic phototrophs, while anaerobic phototrophs are classified as either purple sulfur/nonsulfur bacteria or green sulfur/nonsulfur bacteria. Importantly, it is not clear from surveys of the literature that anaerobic purple bacterial strains (denoted by an asterisk) were robustly tested for their ability to use O2. FNR, ferredoxin-NADP+ oxidoreductase; PFOR, pyruvate-flavodoxin oxidoreductase; Rnf, Rhodobacter nitrogen fixation protein; FeFe, iron-only hydrogenase; NiFe, nickel-iron hydrogenase; Fix, electron transfer flavoprotein involved in nitrogen fixation.
FIG 4
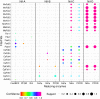
Bubble plot depicting the dominant patterns in the distribution of putative electron carrier protein homologs (Fds/Flds), enzyme homologs that putatively reduce Fd/Fld, and specified Nif lineages, as determined by the Apriori algorithm (124). Each unique pattern is given as a bubble, and the color represents the confidence value, or statistical significance, in the codistribution of specified proteins (only confidence values of ≥0.6 are presented). The size of the bubble represents the support value (≥0.2), or the frequency with which two proteins are identified in the same genome. For simplicity, only the proteins that were present in >20% of the diazotrophic genomes for each specified nitrogenase lineage were considered in this analysis. PFOR, pyruvate-Fld oxidoreductase; Rnf, Rhodobacter nitrogen fixation protein; FeFe, iron-only hydrogenase; NiFe, nickel-iron hydrogenase; FixABCX, electron transfer flavoprotein that is involved in nitrogen fixation. Names of source organisms for Fds and Flds are abbreviated as follows: Mv, Methanocaldococcus vulcanius; Mm, Methanococcus maripaludis; Mb, Methanosarcina barkeri; Cs, Caldicellulosiruptor saccharolyticus; Cp, Clostridium pasteurianum; Av, A. vinelandii. Abbreviations for Fds and Flds are presented in Tables 1 and 2, respectively.
FIG 5
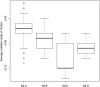
Box plot representing the average oxidation state of carbon in proteomes (inferred from genomes) for the taxa comprising organisms encoding the specified nitrogenase isoform. Here, the box represents the interquartile range and the whiskers show the full range of the data. Outlier values are represented as circles, and the line within the box represents the median value.
Similar articles
- Mechanisms for Generating Low Potential Electrons across the Metabolic Diversity of Nitrogen-Fixing Bacteria.
Alleman AB, Peters JW. Alleman AB, et al. Appl Environ Microbiol. 2023 May 31;89(5):e0037823. doi: 10.1128/aem.00378-23. Epub 2023 May 8. Appl Environ Microbiol. 2023. PMID: 37154716 Free PMC article. Review. - Rnf and Fix Have Specific Roles during Aerobic Nitrogen Fixation in Azotobacter vinelandii.
Alleman AB, Garcia Costas A, Mus F, Peters JW. Alleman AB, et al. Appl Environ Microbiol. 2022 Sep 13;88(17):e0104922. doi: 10.1128/aem.01049-22. Epub 2022 Aug 24. Appl Environ Microbiol. 2022. PMID: 36000884 Free PMC article. - Evolving a New Electron Transfer Pathway for Nitrogen Fixation Uncovers an Electron Bifurcating-Like Enzyme Involved in Anaerobic Aromatic Compound Degradation.
Lewis NM, Sarne A, Fixen KR. Lewis NM, et al. mBio. 2023 Feb 28;14(1):e0288122. doi: 10.1128/mbio.02881-22. Epub 2023 Jan 16. mBio. 2023. PMID: 36645294 Free PMC article. - Evolution of molybdenum nitrogenase during the transition from anaerobic to aerobic metabolism.
Boyd ES, Costas AM, Hamilton TL, Mus F, Peters JW. Boyd ES, et al. J Bacteriol. 2015 May;197(9):1690-9. doi: 10.1128/JB.02611-14. Epub 2015 Mar 2. J Bacteriol. 2015. PMID: 25733617 Free PMC article. - H2 metabolism in photosynthetic bacteria and relationship to N2 fixation.
Willison JC, Jouanneau Y, Colbeau A, Vignais PM. Willison JC, et al. Ann Microbiol (Paris). 1983 Jul-Aug;134B(1):115-35. doi: 10.1016/s0769-2609(83)80100-8. Ann Microbiol (Paris). 1983. PMID: 6139053 Review.
Cited by
- Changes in Carbon Oxidation State of Metagenomes Along Geochemical Redox Gradients.
Dick JM, Yu M, Tan J, Lu A. Dick JM, et al. Front Microbiol. 2019 Feb 11;10:120. doi: 10.3389/fmicb.2019.00120. eCollection 2019. Front Microbiol. 2019. PMID: 30804909 Free PMC article. - Biosynthesis of Nitrogenase Cofactors.
Burén S, Jiménez-Vicente E, Echavarri-Erasun C, Rubio LM. Burén S, et al. Chem Rev. 2020 Jun 24;120(12):4921-4968. doi: 10.1021/acs.chemrev.9b00489. Epub 2020 Jan 24. Chem Rev. 2020. PMID: 31975585 Free PMC article. Review. - Biochemical and Genetic Approaches Improving Nitrogen Use Efficiency in Cereal Crops: A Review.
Sandhu N, Sethi M, Kumar A, Dang D, Singh J, Chhuneja P. Sandhu N, et al. Front Plant Sci. 2021 Jun 4;12:657629. doi: 10.3389/fpls.2021.657629. eCollection 2021. Front Plant Sci. 2021. PMID: 34149755 Free PMC article. Review. - Genome analysis of Pseudomonas sp. OF001 and Rubrivivax sp. A210 suggests multicopper oxidases catalyze manganese oxidation required for cylindrospermopsin transformation.
Martínez-Ruiz EB, Cooper M, Barrero-Canosa J, Haryono MAS, Bessarab I, Williams RBH, Szewzyk U. Martínez-Ruiz EB, et al. BMC Genomics. 2021 Jun 22;22(1):464. doi: 10.1186/s12864-021-07766-0. BMC Genomics. 2021. PMID: 34157973 Free PMC article. - Mechanisms for Generating Low Potential Electrons across the Metabolic Diversity of Nitrogen-Fixing Bacteria.
Alleman AB, Peters JW. Alleman AB, et al. Appl Environ Microbiol. 2023 May 31;89(5):e0037823. doi: 10.1128/aem.00378-23. Epub 2023 May 8. Appl Environ Microbiol. 2023. PMID: 37154716 Free PMC article. Review.
References
- Falkowski PG. 1997. Evolution of the nitrogen cycle and its influence on the biological sequestration of CO2 in the ocean. Nature 387:272–275. doi: 10.1038/387272a0. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical