VIRMA mediates preferential m6A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation - PubMed (original) (raw)
doi: 10.1038/s41421-018-0019-0. eCollection 2018.
Jun Liu 2, Xiaolong Cui 2, Jie Cao 1, Guanzheng Luo 2, Zezhou Zhang 1, Tao Cheng 1, Minsong Gao 1, Xiao Shu 1, Honghui Ma 2, Fengqin Wang 3, Xinxia Wang 3, Bin Shen 4, Yizhen Wang 3, Xinhua Feng 5, Chuan He 2, Jianzhao Liu 1 5
Affiliations
- PMID: 29507755
- PMCID: PMC5826926
- DOI: 10.1038/s41421-018-0019-0
VIRMA mediates preferential m6A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation
Yanan Yue et al. Cell Discov. 2018.
Abstract
_N_6-methyladenosine (m6A) is enriched in 3'untranslated region (3'UTR) and near stop codon of mature polyadenylated mRNAs in mammalian systems and has regulatory roles in eukaryotic mRNA transcriptome switch. Significantly, the mechanism for this modification preference remains unknown, however. Herein we report a characterization of the full m6A methyltransferase complex in HeLa cells identifying METTL3/METTL14/WTAP/VIRMA/HAKAI/ZC3H13 as the key components, and we show that VIRMA mediates preferential mRNA methylation in 3'UTR and near stop codon. Biochemical studies reveal that VIRMA recruits the catalytic core components METTL3/METTL14/WTAP to guide region-selective methylations. Around 60% of VIRMA mRNA immunoprecipitation targets manifest strong m6A enrichment in 3'UTR. Depletions of VIRMA and METTL3 induce 3'UTR lengthening of several hundred mRNAs with over 50% targets in common. VIRMA associates with polyadenylation cleavage factors CPSF5 and CPSF6 in an RNA-dependent manner. Depletion of CPSF5 leads to significant shortening of 3'UTR of over 2800 mRNAs, 84% of which are modified with m6A and have increased m6A peak density in 3'UTR and near stop codon after CPSF5 knockdown. Together, our studies provide insights into m6A deposition specificity in 3'UTR and its correlation with alternative polyadenylation.
Conflict of interest statement
C.H. is a scientific founder of the Accent Therapeutics and a member of the Scientific Advisory Committee. The remaining authors declare that they have no conflict of interest.
Figures
Fig. 1
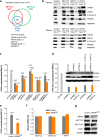
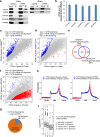
Proteomic identification of new m6A methyltransferase complex components and evaluation of their effects on mRNA m6A modification distribution. a Overlap of protein interactomes of METTL3, METTL14, and WTAP. Stable expression HeLa cells with dual-tagged (N-term tandem Flag and HA) METTL3, METTL14, and WTAP were subjected to tandem affinity purification with Flag and HA antibodies and further mass spectrometry identification. b Validation of selected common targets of METTL3, METTL14, and WTAP by western blotting. After Flag IP in the dual Flag-HA-tagged METTL3, METTL14, and WTAP stable cell lines, western blotting was performed to validate the interactions using endogenous antibodies. Because endogenous ZC3H13 antibody is not good for western blotting, Flag-tagged full-length ZC3H13 and its truncated forms N-terminal (N-ZC3H13) and C-terminal (C-ZC3H13) were overexpressed in the METTL3, METTL14, and WTAP stable cell lines for validation using Flag antibody after HA IP. c Effects of siRNA knockdown of VIRMA, HAKAI, ZC3H13, TRIM28, and HNRNPH on m6A level in HeLa cell line. Two different siRNA constructs were tested for each gene. Data are represented as means ± SEM of n = 3. P values calculated using one-tailed Student’s _t_-test between control and siRNA knockdown samples are <0.05, except for TRIM28 knockdown samples (_P_ > 0.05). d Effects of VIRMA siRNA knockdowns on WTAP expression and polyadenylated RNA m6A levels. Overexpression (oe) of WTAP under VIRMA siRNA knockdowns cannot recover m6A level. Data are represented as means ± SEM of n = 3. P values calculated using one-tailed Student’s _t_-test between control and siRNA knockdown samples are <0.001, while _P_ values between siVIRMA and siVIRMA/WTAP oe are larger than 0.05. **e** Comparison of m6A levels in polyadenylated RNAs in between control and _VIRMA_mut/− HeLa cell lines. The _VIRMA_mut/− HeLa cell line was generated by cas9 gene-editing system. Data are represented as means ± SEM of _n_ = 3. _P_ values calculated using one-tailed Student’s _t_-test between control and _VIRMA_mut/− samples are <0.001. **f**, **g** Comparison of mRNA and protein expression levels of METTL3, METTL14, and WTAP between control and _VIRMA_mut/− HeLa cell lines using RT-qPCR (**f**) and western blotting (**g**), respectively. GAPDH serves as control. _P_ values calculated using one-tailed Student’s _t_-test between control and _VIRMA_mut/− RT-qPCR samples are >0.05
Fig. 2
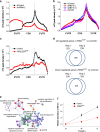
VIRMA affects cellular functions. a, b Effects of depletion of VIRMA (a) and METTL3, METTL14, and WTAP (b) by siRNA knockdowns on the profiles of m6A peak density along mRNA transcript. c Effect of partial VIRMA knockout (_VIRMA_mut/−) on the profiles of m6A peak density along mRNA transcript. d Differentially expressed genes (fold change > 2) were identified by the RNA-seq in between _VIRMA_mut/− and control cell lines. The number of overlapped upregulated and downregulated genes are shown. e Gene ontology (GO) and enrichment analysis of the differentially expressed and m6A-containing genes. f Estimate of VIRMA depletion on cell proliferation by MTS assay. Data are represented as means ± SEM of n = 3
Fig. 3
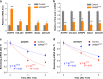
VIRMA modulates abundance and lifetime of mRNAs through mediating m6A installation. a,b Validation of selected mRNA targets with upregulated expression (a) and decreased m6A level (b) upon VIRMA depletion in HeLa cell line. P values calculated using one-tailed Student’s _t_-test between control and VIRMA depletion samples; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Data are represented as means ± SEM of n = 3. c, d Measurement of lifetimes of selected mRNAs IGFBP5 (c) and NOTCH1 (d). Data are represented as means ± SEM of n = 3
Fig. 4
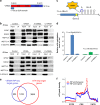
VIRMA recruits catalytic components METTL3/METTL14/WTAP in order to guide mRNA m6A methylation at specific site. a Schematic of domain architecture (aa, amino acids) of VIRMA, N terminus of VIRMA (N-VIRMA, aa 1–1130), and C terminus of VIRMA (C-VIRMA, aa 1131–1812). b Co-immunoprecipitation experiments in HeLa cells in order to dissect interactions of different domains of VIRMA with other methyltransferase components. RNase was added in order to determine if the interaction was RNA-dependent. HSV-tagged C-ZC3H13 was co-expressed for immunoprecipitation experiment and corresponding HSV-tag antibody was used in the western blotting. c Construct of the tethering reporter assay. The mRNA reporter consists of a firefly luciferase sequence as the coding region and five Box B sequence at 3′UTR (F-Luc-5BoxB). There exists a GGACU motif near the stop codon. Both N-VIRMA and C-VIRMA were fused with λ peptide (N-VIRMA-λ and C-VIRMA-λ), which recognizes Box B RNA with a high affinity. d Measurement of m6A level of F-Luc-5BoxB mRNAs under co-expression with different truncated forms of VIRMA. The synonymously mutated construct with GGAUU was tested for comparison. Renilla luciferase was used as an internal control to normalize the F-Luc signal. e Overlap of VIRMA RIP-seq mRNA targets with m6A-seq targets in HeLa cells. Endogenous VIRMA antibody was used for direct RNA immunoprecipitation. RIP-seq-enriched genes were defined as FDR ≤ 0.05 and log2(IP/Input) ≥ 1. f Comparison of m6A peak density profiles between RIP-seq-enriched and control genes. RIP-seq control genes (1909) were defined as FDR ≤ 0.05 and log2(IP/Input) < 1
Fig. 5
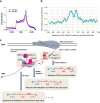
VIRMA associates with polyadenylation cleavage factors. a Western blots to validate potential proteomic targets of VIRMA, including polyadenylation cleavage factors CPSF5 and CPSF6, cleavage stimulation factor CSTF2T, poly(A)-binding protein PABP1, RNA polymerase II subunit POLR2B, and transcriptional intermediary factor TRIM28. b Effect of siRNA knockdown of CPSF5 and CPSF6 on m6A level of polyadenylated RNAs. P values calculated using one-tailed Student’s _t_-test between control and knockdown samples are >0.05. Data are represented as means ± SEM of n = 3. c, d Scattering plots of PDUIs in between control and VIRMA (c) and METTL3 (d) mutant cells, where 3′UTRs of mRNAs are significantly shortened or lengthened with FDR ≤ 0.05, absolute ΔPDUI ≥ 0.2, and at least twofold change of PUDIs. e Overlap of 3′UTR-lengthened genes regulated by VIRMA and METTL3 depletion effects. f Scattering plots of PDUIs in between siControl and siCPSF5 cells, where 3′UTRs of mRNAs are significantly shortened or lengthened with FDR ≤ 0.05, absolute ΔPDUI ≥ 0.2, and at least twofold change of PUDIs. g Pie chart shows percentages of m6A and non-m6A genes with 3′UTR shortening under the CPSF5 knockdown. Eighty-four percent of shortened genes are enriched with m6A modification. Chi-square test gives a P value < 1e-16. h The m6A peak density profiles of m6A genes in HeLa siControl and siCPSF5 cells grouped by their 3′UTR shortening induced by CPSF5 knockdown. i Differential gene expressions for 3′UTR-shortened m6A and non-m6A genes, respectively, after CPSF5 knockdown. RPKM stands for reads per kilobase per million mapped reads
Fig. 6
Correlation of 3′UTR m6A methylation with alternative polyadenylation. a The m6A peak density profiles of m6A genes in HeLa siControl and siCPSF5 cells. b m6A GGACU motif coverage versus motif distance from proximal APA site along mRNA transcript. c Proposed model for VIRMA-regulated m6A methylation specificity in 3′UTR and near stop codon and correlation of m6A methylation with APA
Similar articles
- Wilms' tumor 1-associating protein complex regulates alternative splicing and polyadenylation at potential G-quadruplex-forming splice site sequences.
Horiuchi K, Kawamura T, Hamakubo T. Horiuchi K, et al. J Biol Chem. 2021 Nov;297(5):101248. doi: 10.1016/j.jbc.2021.101248. Epub 2021 Sep 25. J Biol Chem. 2021. PMID: 34582888 Free PMC article. - The drive to generate multiple forms of oncogenic cyclin D1 transcripts in mantle cell lymphoma.
Masamha CP. Masamha CP. Biomark Res. 2017 May 8;5:16. doi: 10.1186/s40364-017-0094-7. eCollection 2017. Biomark Res. 2017. PMID: 28503306 Free PMC article. - Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control.
Li W, Park JY, Zheng D, Hoque M, Yehia G, Tian B. Li W, et al. BMC Biol. 2016 Jan 22;14:6. doi: 10.1186/s12915-016-0229-6. BMC Biol. 2016. PMID: 26801249 Free PMC article. - A birds'-eye view of the activity and specificity of the mRNA m6 A methyltransferase complex.
Garcias Morales D, Reyes JL. Garcias Morales D, et al. Wiley Interdiscip Rev RNA. 2021 Jan;12(1):e1618. doi: 10.1002/wrna.1618. Epub 2020 Jul 19. Wiley Interdiscip Rev RNA. 2021. PMID: 32686365 Review. - Structures and mechanisms of the RNA m 6A writer.
Deng T, Ma J. Deng T, et al. Acta Biochim Biophys Sin (Shanghai). 2024 Sep 4;57(1):59-72. doi: 10.3724/abbs.2024152. Acta Biochim Biophys Sin (Shanghai). 2024. PMID: 39238441 Free PMC article. Review.
Cited by
- Decoding the epitranscriptome: a new frontier for cancer therapy and drug resistance.
Tang L, Tian H, Min Q, You H, Yin M, Yang L, Zhao Y, Wu X, Li M, Du F, Chen Y, Deng S, Li X, Chen M, Gu L, Sun Y, Xiao Z, Li W, Shen J. Tang L, et al. Cell Commun Signal. 2024 Oct 21;22(1):513. doi: 10.1186/s12964-024-01854-w. Cell Commun Signal. 2024. PMID: 39434167 Free PMC article. Review. - Dual effects of N6-methyladenosine on cancer progression and immunotherapy.
Li H, Wu H, Wang Q, Ning S, Xu S, Pang D. Li H, et al. Mol Ther Nucleic Acids. 2021 Feb 10;24:25-39. doi: 10.1016/j.omtn.2021.02.001. eCollection 2021 Jun 4. Mol Ther Nucleic Acids. 2021. PMID: 33738136 Free PMC article. Review. - METTL16, Methyltransferase-Like Protein 16: Current Insights into Structure and Function.
Ruszkowska A. Ruszkowska A. Int J Mol Sci. 2021 Feb 22;22(4):2176. doi: 10.3390/ijms22042176. Int J Mol Sci. 2021. PMID: 33671635 Free PMC article. Review. - m6A methylation modification and immune cell infiltration: implications for targeting the catalytic subunit m6A-METTL complex in gastrointestinal cancer immunotherapy.
Peng C, Xiong F, Pu X, Hu Z, Yang Y, Qiao X, Jiang Y, Han M, Wang D, Li X. Peng C, et al. Front Immunol. 2023 Dec 15;14:1326031. doi: 10.3389/fimmu.2023.1326031. eCollection 2023. Front Immunol. 2023. PMID: 38187373 Free PMC article. Review. - Identification and validation of signature for prognosis and immune microenvironment in gastric cancer based on m6A demethylase ALKBH5.
Ji T, Gao X, Li D, Huai S, Chi Y, An X, Ji W, Yang S, Li J. Ji T, et al. Front Oncol. 2023 Jan 6;12:1079402. doi: 10.3389/fonc.2022.1079402. eCollection 2022. Front Oncol. 2023. PMID: 36686788 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases