Replication Study: Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma - PubMed (original) (raw)
Meta-Analysis
Replication Study: Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma
John Repass et al. Elife. 2018.
Abstract
As part of the Reproducibility Project: Cancer Biology, we published a Registered Report (Repass et al., 2016), that described how we intended to replicate an experiment from the paper 'Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma' (Castellarin et al., 2012). Here we report the results. When measuring Fusobacterium nucleatum DNA by qPCR in colorectal carcinoma (CRC), adjacent normal tissue, and separate matched control tissue, we did not detect a signal for F. nucleatum in most samples: 25% of CRCs, 15% of adjacent normal, and 0% of matched control tissue were positive based on quantitative PCR (qPCR) and confirmed by sequencing of the qPCR products. When only samples with detectable F. nucleatum in CRC and adjacent normal tissue were compared, the difference was not statistically significant, while the original study reported a statistically significant increase in F. nucleatum expression in CRC compared to adjacent normal tissue (Figure 2; Castellarin et al., 2012). Finally, we report a meta-analysis of the result, which suggests F. nucleatum expression is increased in CRC, but is confounded by the inability to detect F. nucleatum in most samples. The difference in F. nucleatum expression between CRC and adjacent normal tissues was thus smaller than the original study, and not detected in most samples.
Keywords: Fusobacterium nucleatum; Reproducibility Project: Cancer Biology; cancer biology; colorectal carcinoma; human; metascience; replication; reproducibility.
© 2018, Repass et al.
Conflict of interest statement
JR ARQ Genetics is a Science Exchange associated lab. EI, NP: Employed by and hold shares in Science Exchange Inc.
Figures
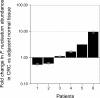
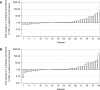
Figure 1.. Relative abundance of F. nucleatum in colorectal carcinoma versus adjacent normal biopsies in samples with detectable F. nucleatum.
Only samples in which F. nucleatum was detected in both colorectal carcinoma (CRC) and adjacent normal tissue by quantitative PCR (qPCR) and confirmed by sequencing of the qPCR product are shown (n = 6). The mean relative abundance of F. nucleatum (normalized to PGT expression) in CRC tissue versus adjacent normal tissue of both independent runs is reported for each patient sample and error bars represent SEM. The y-axis represents mean fold gene expression change (2-ΔΔCt) while the x-axis represents patient samples. Exploratory analysis: two-tailed paired t test: t(5) = 1.067, p = 0.335, r = 0.43, 95% CI [−0.59, 0.92]. Additional details for this experiment can be found at
.
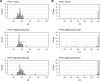
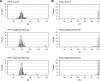
Figure 1—figure supplement 1.. First independent qPCR run.
First of two independent qPCR runs performed in technical duplicate. (A) Distribution of raw Ct values for PGT in colorectal carcinoma (CRC) tumor, adjacent normal tissue, or matched normal samples. (B) Distribution of raw Ct values for F. nucleatum in each sample type (CRC tumor, adjacent normal tissue, or matched normal). The y-axis represents a binned count of each Ct score and the x-axis represents the value of Ct scores. Bin size = 0.1. Additional details can be found at
.
Figure 1—figure supplement 2.. Second independent qPCR run.
Second of two independent qPCR runs performed in technical duplicate. (A) Distribution of raw Ct values for PGT in colorectal carcinoma (CRC) tumor, adjacent normal tissue, or matched normal samples. (B) Distribution of raw Ct values for F. nucleatum in each sample type (CRC tumor, adjacent normal tissue, or matched normal). The y-axis represents a binned count of each Ct score and the x-axis represents the value of Ct scores. Bin size = 0.1. Additional details can be found at
.
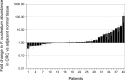
Figure 2.. Relative abundance of F. nucleatum by qPCR in colorectal carcinoma versus adjacent normal biopsies.
Fold change values are shown for all paired specimens based on differences in Ct values, irrespective of whether the qPCR products were confirmed to be specific or non-specific upon sequencing. Tissue was collected from colorectal carcinoma (CRC) and adjacent normal tissue (n = 40). qPCR was performed independently two times and averaged. The mean relative abundance of F. nucleatum (normalized to PGT expression) in CRC tissue versus adjacent normal tissue of both independent runs is reported for each patient sample and error bars represent SEM. The y-axis represents mean fold gene expression change (2-ΔΔCt) while the x-axis represents patient samples. Two-tailed Wilcoxon signed-rank test: Z = 2.14, p = 0.032. Additional details for this experiment can be found at
.
Figure 2—figure supplement 1.. Independent qPCR runs.
Fold change values are shown for all paired specimens based on differences in Ct values, irrespective of whether the qPCR products were confirmed to be specific or non-specific upon sequencing. (A) First of two independent qPCR runs performed in technical duplicate. Mean relative abundance of F. nucleatum (normalized to PGT expression) in colorectal carcinoma (CRC) tissue versus adjacent normal tissue is reported for each patient sample. The y-axis represents mean fold gene expression change (2-ΔΔCt) and the x-axis represents patient samples. The median fold change across all samples was 1.11 (IQR = 0.89–2.18) and the mean fold change was 3.62 [SD = 7.21]. (B) Second of two independent qPCR runs performed in technical duplicate. Mean relative abundance of F. nucleatum (normalized to PGT expression) in CRC tissue versus adjacent normal tissue is reported for each patient sample. The y-axis represents mean fold gene expression change (2-ΔΔCt) and the x-axis represents patient samples. The median fold change across all samples was 1.13 (IQR = 0.90–2.54) and the mean fold change was 8.27 [SD = 32.2]. Additional details can be found at
.
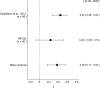
Figure 3.. Meta-analyses of each effect.
Effect size (r) and 95% confidence interval are presented for Castellarin et al. (2012), this replication attempt (RP:CB), and a meta-analysis to combine the two effects. To directly compare and combine the results of both studies, the qPCR results for RP:CB were used irrespective of whether the amplicon was confirmed to be F. nucleatum by sequencing. The effect size r is a standardized measure of the correlation (strength and direction) of the association between tissue type and normalized F. nucleatum DNA levels, with a larger positive value indicating CRC tissue is correlated with a higher F. nucleatum expression level. Sample sizes used in Castellarin et al. (2012) and this replication attempt are reported under the study name. Random effects meta-analysis of the abundance of F. nucleatum in CRC tissue compared to adjacent normal tissue (meta-analysis p = 5.86 × 10−4). Additional details for this meta-analysis can be found at
.
Comment in
- The who, where and how of fusobacteria and colon cancer.
Sears CL. Sears CL. Elife. 2018 Mar 13;7:e28434. doi: 10.7554/eLife.28434. Elife. 2018. PMID: 29533185 Free PMC article.
Similar articles
- Registered report: Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma.
Repass J, Maherali N, Owen K; Reproducibility Project: Cancer Biology; Reproducibility Project Cancer Biology. Repass J, et al. Elife. 2016 Feb 11;5:e10012. doi: 10.7554/eLife.10012. Elife. 2016. PMID: 26882501 Free PMC article. - Association Between Fusobacterium nucleatum, Pathway Mutation, and Patient Prognosis in Colorectal Cancer.
Lee DW, Han SW, Kang JK, Bae JM, Kim HP, Won JK, Jeong SY, Park KJ, Kang GH, Kim TY. Lee DW, et al. Ann Surg Oncol. 2018 Oct;25(11):3389-3395. doi: 10.1245/s10434-018-6681-5. Epub 2018 Jul 30. Ann Surg Oncol. 2018. PMID: 30062471 - Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population.
Yamaoka Y, Suehiro Y, Hashimoto S, Hoshida T, Fujimoto M, Watanabe M, Imanaga D, Sakai K, Matsumoto T, Nishioka M, Takami T, Suzuki N, Hazama S, Nagano H, Sakaida I, Yamasaki T. Yamaoka Y, et al. J Gastroenterol. 2018 Apr;53(4):517-524. doi: 10.1007/s00535-017-1382-6. Epub 2017 Aug 19. J Gastroenterol. 2018. PMID: 28823057 - Fusobacterium's link to colorectal neoplasia sequenced: A systematic review and future insights.
Hussan H, Clinton SK, Roberts K, Bailey MT. Hussan H, et al. World J Gastroenterol. 2017 Dec 28;23(48):8626-8650. doi: 10.3748/wjg.v23.i48.8626. World J Gastroenterol. 2017. PMID: 29358871 Free PMC article. Review. - Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer.
Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H, Maruyama R, Imai K, Yamamoto H, Shinomura Y. Nosho K, et al. World J Gastroenterol. 2016 Jan 14;22(2):557-66. doi: 10.3748/wjg.v22.i2.557. World J Gastroenterol. 2016. PMID: 26811607 Free PMC article. Review.
Cited by
- Comparative Analysis of Colon Cancer-Derived Fusobacterium nucleatum Subspecies: Inflammation and Colon Tumorigenesis in Murine Models.
Queen J, Domingue JC, White JR, Stevens C, Udayasuryan B, Nguyen TTD, Wu S, Ding H, Fan H, McMann M, Corona A, Larman TC, Verbridge SS, Housseau F, Slade DJ, Drewes JL, Sears CL. Queen J, et al. mBio. 2021 Feb 22;13(1):e0299121. doi: 10.1128/mbio.02991-21. Epub 2022 Feb 8. mBio. 2021. PMID: 35130731 Free PMC article. - Understanding of researcher behavior is required to improve data reliability.
Wass MN, Ray L, Michaelis M. Wass MN, et al. Gigascience. 2019 May 1;8(5):giz017. doi: 10.1093/gigascience/giz017. Gigascience. 2019. PMID: 30715291 Free PMC article. - Intratumoural microbiota: a new frontier in cancer development and therapy.
Cao Y, Xia H, Tan X, Shi C, Ma Y, Meng D, Zhou M, Lv Z, Wang S, Jin Y. Cao Y, et al. Signal Transduct Target Ther. 2024 Jan 10;9(1):15. doi: 10.1038/s41392-023-01693-0. Signal Transduct Target Ther. 2024. PMID: 38195689 Free PMC article. Review. - Colorectal cancer: genetic abnormalities, tumor progression, tumor heterogeneity, clonal evolution and tumor-initiating cells.
Testa U, Pelosi E, Castelli G. Testa U, et al. Med Sci (Basel). 2018 Apr 13;6(2):31. doi: 10.3390/medsci6020031. Med Sci (Basel). 2018. PMID: 29652830 Free PMC article. Review. - Fusobacterium nucleatum promotes inflammatory and anti-apoptotic responses in colorectal cancer cells via ADP-heptose release and ALPK1/TIFA axis activation.
Martin-Gallausiaux C, Salesse L, Garcia-Weber D, Marinelli L, Beguet-Crespel F, Brochard V, Le Gléau C, Jamet A, Doré J, Blottière HM, Arrieumerlou C, Lapaque N. Martin-Gallausiaux C, et al. Gut Microbes. 2024 Jan-Dec;16(1):2295384. doi: 10.1080/19490976.2023.2295384. Epub 2023 Dec 21. Gut Microbes. 2024. PMID: 38126163 Free PMC article.
References
- Bradley WH, Eng K, Le M, Mackinnon AC, Kendziorski C, Rader JS. Comparing gene expression data from formalin-fixed, paraffin embedded tissues and qPCR with that from snap-frozen tissue and microarrays for modeling outcomes of patients with ovarian carcinoma. BMC Clinical Pathology. 2015;15:17. doi: 10.1186/s12907-015-0017-1. - DOI - PMC - PubMed
- Caraguel CG, Stryhn H, Gagné N, Dohoo IR, Hammell KL. Selection of a cutoff value for real-time polymerase chain reaction results to fit a diagnostic purpose: analytical and epidemiologic approaches. Journal of Veterinary Diagnostic Investigation. 2011;23:2–15. doi: 10.1177/104063871102300102. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
The funder had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials