The global distribution and spread of the mobilized colistin resistance gene mcr-1 - PubMed (original) (raw)
doi: 10.1038/s41467-018-03205-z.
Lucy van Dorp 2, Liam P Shaw 2, Phelim Bradley 3, Qi Wang 1, Xiaojuan Wang 1, Longyang Jin 1, Qing Zhang 4, Yuqing Liu 4, Adrien Rieux 5, Thamarai Dorai-Schneiders 6, Lucy Anne Weinert 7, Zamin Iqbal 3 8, Xavier Didelot 9, Hui Wang 10, Francois Balloux 11
Affiliations
- PMID: 29563494
- PMCID: PMC5862964
- DOI: 10.1038/s41467-018-03205-z
The global distribution and spread of the mobilized colistin resistance gene mcr-1
Ruobing Wang et al. Nat Commun. 2018.
Abstract
Colistin represents one of the few available drugs for treating infections caused by carbapenem-resistant Enterobacteriaceae. As such, the recent plasmid-mediated spread of the colistin resistance gene mcr-1 poses a significant public health threat, requiring global monitoring and surveillance. Here, we characterize the global distribution of mcr-1 using a data set of 457 mcr-1-positive sequenced isolates. We find mcr-1 in various plasmid types but identify an immediate background common to all mcr-1 sequences. Our analyses establish that all mcr-1 elements in circulation descend from the same initial mobilization of mcr-1 by an ISApl1 transposon in the mid 2000s (2002-2008; 95% highest posterior density), followed by a marked demographic expansion, which led to its current global distribution. Our results provide the first systematic phylogenetic analysis of the origin and spread of mcr-1, and emphasize the importance of understanding the movement of antibiotic resistance genes across multiple levels of genomic organization.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Fig. 1
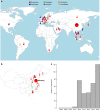
Overview of the _mcr-1-_positive isolates included. a Global map of _mcr-1-_positive isolates included colored by genus with the number and size of pies providing the sample size per location; b Map of novel Chinese isolates sequenced for this study; c Histogram of sampling dates (years) of the isolates. Maps were created using the R package rworldmap using the public domain Natural Earth data set
Fig. 2
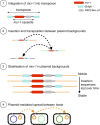
Schematic representation of the evolutionary model for the steps in the spread of the mcr-1 gene. (1) The formation of the original composite transposon, followed by (2) transposition between plasmid backgrounds and (3) stabilization via loss of IS_A_ pl1 elements before (4) plasmid-mediated spread
Fig. 3
The genetic element carrying mcr-1 is a composite transposon and is alignable across our global data set. a Length distribution of the alignment across sequences. b Length distribution subset by plasmid type. c The composite transposon, consisting of IS_Apl_ 1, mcr-1, a PAP2 orf, and IS_Apl_ 1. The region indicated by the red arrow was used in phylogenetic analyses, after the removal of recombination. d The 186 bp region upstream of mcr-1 showed strong signals of recombination (gray box) that coincided with the promotor regions of mcr-1 (red box), and this diverse region was removed from the subsequent alignment. e Twenty-eight sequences from Vietnam had a 1.7 kb insertion containing a putative transpose, suggesting subsequent rearrangement after initial mobilization
Fig. 4
Phylogeny of the mcr-1 composite transposon indicates a dominant sequence type with subsequent diversification. Midpoint-rooted maximum parsimony phylogeny based on the 3522 bp alignment of 457 sequences (recombinant regions removed). Size of points indicates the number of identical sequences, with a representative sequence for each shown next to each tip
Fig. 5
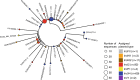
The distribution of plasmid types shown on the transposon phylogeny. Maximum parsimony tree (homoplastic sites removed, midpoint rooted, as in Fig. 4) based on the composite transposon alignment for 172 sequences containing a plasmid replicon on the same contig i.e. those with an assigned plasmid type (color). IncI2 and IncX4 are the most common plasmid types. An example sequence ID is shown for each unique sequence
Similar articles
- Microbiological surveillance of plasmid mediated colistin resistance in human Enterobacteriaceae isolates in Romagna (Northern Italy): August 2016-July 2017.
Del Bianco F, Morotti M, Pedna MF, Farabegoli P, Sambri V. Del Bianco F, et al. Int J Infect Dis. 2018 Apr;69:96-98. doi: 10.1016/j.ijid.2018.02.006. Epub 2018 Feb 12. Int J Infect Dis. 2018. PMID: 29447913 - High Rates of Human Fecal Carriage of mcr-1-Positive Multidrug-Resistant Enterobacteriaceae Emerge in China in Association With Successful Plasmid Families.
Zhong LL, Phan HTT, Shen C, Vihta KD, Sheppard AE, Huang X, Zeng KJ, Li HY, Zhang XF, Patil S, Crook DW, Walker AS, Xing Y, Lin JL, Feng LQ, Doi Y, Xia Y, Stoesser N, Tian GB. Zhong LL, et al. Clin Infect Dis. 2018 Feb 10;66(5):676-685. doi: 10.1093/cid/cix885. Clin Infect Dis. 2018. PMID: 29040419 Free PMC article. - Detection of mcr-1 Plasmids in Enterobacteriaceae Isolates From Human Specimens: Comparison With Those in Escherichia coli Isolates From Livestock in Korea.
Yoon EJ, Hong JS, Yang JW, Lee KJ, Lee H, Jeong SH. Yoon EJ, et al. Ann Lab Med. 2018 Nov;38(6):555-562. doi: 10.3343/alm.2018.38.6.555. Ann Lab Med. 2018. PMID: 30027699 Free PMC article. - Transferable resistance to colistin: a new but old threat.
Schwarz S, Johnson AP. Schwarz S, et al. J Antimicrob Chemother. 2016 Aug;71(8):2066-70. doi: 10.1093/jac/dkw274. Epub 2016 Jun 24. J Antimicrob Chemother. 2016. PMID: 27342545 Review. - Colistin use and colistin resistance in bacteria from animals.
Kempf I, Jouy E, Chauvin C. Kempf I, et al. Int J Antimicrob Agents. 2016 Dec;48(6):598-606. doi: 10.1016/j.ijantimicag.2016.09.016. Epub 2016 Oct 27. Int J Antimicrob Agents. 2016. PMID: 27836380 Review.
Cited by
- Destination shapes antibiotic resistance gene acquisitions, abundance increases, and diversity changes in Dutch travelers.
D'Souza AW, Boolchandani M, Patel S, Galazzo G, van Hattem JM, Arcilla MS, Melles DC, de Jong MD, Schultsz C; COMBAT Consortium; Dantas G, Penders J. D'Souza AW, et al. Genome Med. 2021 Jun 7;13(1):79. doi: 10.1186/s13073-021-00893-z. Genome Med. 2021. PMID: 34092249 Free PMC article. - Genome Characterization of _mcr-1_-Positive Escherichia coli Isolated From Pigs With Postweaning Diarrhea in China.
Guo L, Wang J, Wang S, Su J, Wang X, Zhu Y. Guo L, et al. Front Vet Sci. 2020 Aug 25;7:503. doi: 10.3389/fvets.2020.00503. eCollection 2020. Front Vet Sci. 2020. PMID: 33005637 Free PMC article. - A Large Spatial Survey of Colistin-Resistant Gene _mcr-1_-Carrying E. coli in Rivers across Taiwan.
Teng CH, Wu PC, Tang SL, Chen YC, Cheng MF, Huang PC, Ko WC, Wang JL. Teng CH, et al. Microorganisms. 2021 Mar 31;9(4):722. doi: 10.3390/microorganisms9040722. Microorganisms. 2021. PMID: 33807253 Free PMC article. - Global Variation in Escherichia coli mcr-1 Genes and Plasmids from Animal and Human Genomes Following Colistin Usage Restrictions in Livestock.
Garcias B, Flores MA, Fernández M, Monteith W, Pascoe B, Sheppard SK, Martín M, Cortey M, Darwich L. Garcias B, et al. Antibiotics (Basel). 2024 Aug 12;13(8):759. doi: 10.3390/antibiotics13080759. Antibiotics (Basel). 2024. PMID: 39200059 Free PMC article. - Inter-plasmid transfer of antibiotic resistance genes accelerates antibiotic resistance in bacterial pathogens.
Wang X, Zhang H, Yu S, Li D, Gillings MR, Ren H, Mao D, Guo J, Luo Y. Wang X, et al. ISME J. 2024 Jan 8;18(1):wrad032. doi: 10.1093/ismejo/wrad032. ISME J. 2024. PMID: 38366209 Free PMC article.
References
- Grégoire, N., Aranzana-Climent, V., Magréault, S., Marchand, S. & Couet, W. Clinical pharmacokinetics and pharmacodynamics of colistin. Clin. Pharmacokinet.56 1441–1460 (2017). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials