The kinetoplastid-infecting Bodo saltans virus (BsV), a window into the most abundant giant viruses in the sea - PubMed (original) (raw)
The kinetoplastid-infecting Bodo saltans virus (BsV), a window into the most abundant giant viruses in the sea
Christoph M Deeg et al. Elife. 2018.
Abstract
Giant viruses are ecologically important players in aquatic ecosystems that have challenged concepts of what constitutes a virus. Herein, we present the giant Bodo saltans virus (BsV), the first characterized representative of the most abundant group of giant viruses in ocean metagenomes, and the first isolate of a klosneuvirus, a subgroup of the Mimiviridae proposed from metagenomic data. BsV infects an ecologically important microzooplankton, the kinetoplastid Bodo saltans. Its 1.39 Mb genome encodes 1227 predicted ORFs, including a complex replication machinery. Yet, much of its translational apparatus has been lost, including all tRNAs. Essential genes are invaded by homing endonuclease-encoding self-splicing introns that may defend against competing viruses. Putative anti-host factors show extensive gene duplication via a genomic accordion indicating an ongoing evolutionary arms race and highlighting the rapid evolution and genomic plasticity that has led to genome gigantism and the enigma that is giant viruses.
Keywords: Bodo saltans; Bodo saltans virus; Giant Viruses; NCLDV; evolutionary biology; genomics; infectious disease; microbiology.
© 2018, Deeg et al.
Conflict of interest statement
CD, CC, CS No competing interests declared
Figures
Figure 1.. BsV induced lysis observed by flow cytometry.
(A) Flow cytometry profile of uninfected Bodo saltans stained with Lysotracker (blue arrow head) (B) Flow cytometry profile of BsV (red arrow head) and bacteria (green arrow head) stained with SYBR Green. (C) The abundances of B. saltans cells and BsV particles after infection at a particle to cell ratio of 2.
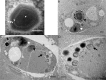
Figure 2.. Ultrastructure of BsV particles and replication.
(A) Mature BsV virion: DNA containing core is surrounded by two putative membranous layers. The capsid consists of at least two proteinaceous layers. The bright halo hints to the presence of short (~40 nm) fibers as observed in ApMV. The top vertex of the virion contains a possible stargate structure (See also Figure 2—figure supplement 1). (Scale bar = 100 nm) (B) Healthy Bodo saltans cell: Nucleus with nucleolus and heterochromatin structures (Back arrow head) and kinetoplast genome (white arrow head) are clearly visible. (Scale bar = 500 nm) (C) Cell of Bodo saltans 24 hr post-BsV infection: Most subcellular compartments of healthy cells have been displaced by the virus factory now taking up a third of the cell. Virion production is directed toward the periphery of the cell (black arrow). Kinetoplast genome remains intact (white arrow head) while the nuclear genome is degraded (black arrow head; Scale bar = 500 nm) (D) BsV virion assembly and maturation: Lipid vesicles migrate through the virion factory where capsid proteins attach for the proteinaceous shell. Vesicles burst and accumulate at the virus factory periphery where the capsid assembly completes (black arrow). Once the capsid is assembled, the virion is filled with the genome and detaches from the virus factory. Internal structures develop inside the virion in the cell’s periphery where mature virions accumulate until the host cell bursts (Scale bar = 500 nm). See Figure 2—figure supplement 1 for further information.
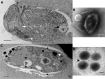
Figure 2—figure supplement 1.. Ultrastructure of BsV particles and replication.
(A) Healthy Bodo saltans cell: Visible structures include the cytostome (black arrow head), the Golgi, mitochondrial arms protruding from the kinetoplast center with the kinetoplast genome (white arrow head), the flagellar root of both flagella as well as several vacuoles (back arrow: food vacuole containing partially digested bacterial prey) are visible traveling from the cytopharynx (white arrow) to the posterior cell pole (Scale bar = 500 nm) (B) Negative staining of a BsV particle with a blossom like opened stargate (C) Bodo saltans cell 24 hr post-BsV infection showing degraded intracellular structures and an extensive BsV virion factory (black arrow head). (D) BsV virions inside a vesicle. A closed stargate is visible at the apex of the bottom left virion (black arrow head).
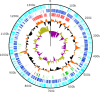
Figure 3.. Circularized genome plot of the linear BsV genome.
Circles from inside out: Olive/Purple: GC-skew; Orange/Black: % GC content plotted around average of 25.3%; Red: Ankyrin repeat domain-containing proteins; Green: Essential NCLDV conserved genes; Dark blue: Plus strand encoded ORFs; Light blue: Negative strand encoded ORFs. See Figure 3—figure supplement 1 for further information on repeat regions.
Figure 3—figure supplement 1.. Ankyrin repeat domain-containing proteins.
(A) Location of recently duplicated region in the 3’ end of the genome (highlighted in black bars, duplicated CDS in red) (B) Alignment of recently duplicated sequence from A, encoding several Ankyrin repeat domain-containing proteins. A region containing four CDS was duplicated in a reverse complement orientation maintaining the integrity and sequence identity of the original sequence (% DNA ID in the central region is 99.7%) (C) ID, annotation and functional class of ankyrin repeat domain-containing proteins with a recognizable domain fragment in the N-terminal region of the protein.
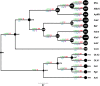
Figure 4.. BsV genome content.
(A) Functional assignment of BsV genome content based on BLASTp and CDD rps-BLAST (B) Domain of best BLASTp hits (C) Evolutionary history of translational machinery found in giant viruses inferred by COUNT. The size of the black circles mapped on a cladogram of the large members of the Mimiviridae (see Figure 6 for full phylogenetic analysis) represents the number of protein coding gene families involved in translation at each node or tip. Blue circles indicate the number of tRNA genes found in each genome. Gene gain and loss events are depicted along the branches. Genomes based on metagenomic assemblies are highlighted to indicate the possibility of incomplete representation of the translation machinery. See Figure 4—figure supplement 3 for the complete phylogenetic tree for members of the Mimiviridae. See Figure 4—figure supplement 2 for a table of all genes included in the analysis. See Figure 4—figure supplement 1 for a cladogram depicting the inferred evolutionary history of all gene families in within the Mimiviridae. MVc: Megavirus chilensis, AMoV: Acanthamoeba polyphaga Moumouvirus, ApMV: Acanthamoeba polyphage Mimivirus, CatV: Catovirus, BsV: Bodo saltans virus, HokV: Hokovirus, KloV: Klosneuvirus (KlosnV), IndV: Indivirus, CroV: Cafeteria roenbergensis virus.
Figure 4—figure supplement 1.. Evolutionary history of gene family content found in giant viruses inferred by COUNT.
The size of the black circles mapped on a cladogram of the Mimiviridae (see Figure 6 for full phylogenetic analysis) represents the number of gene families at each node or tip. Gene gain and loss events are depicted on top of the branches in red and green, gene duplication and contraction in blue and purple below. MVc: Megavirus chilensis, AMoV: Acanthamoeba polyphaga Moumouvirus, ApMV: Acanthamoeba polyphage Mimivirus, CatV: Catovirus, BsV: Bodo saltans virus, HokV: Hokovirus, KloV: Klosneuvirus (KlosnV), IndV: Indivirus, CroV: Cafeteria roenbergensis virus, OLV1: Organic Lake Phycodnavirus 1, OLV2: Organic Lake Phycodnavirus 2, CeV: Chrysochromulina Ericina Virus, PgV: Phaeocystis globosa virus PgV-16T, AaV: Aureococcus anophagefferens virus.
Figure 4—figure supplement 2.. Comparison of translational machinery encoded by giant viruses based on phylogenetic analysis: BsV Translational machinery compared to other NCLDV: blue: monophyletic group, red: recently host acquired, green: recently acquired from bodonid host, ψ: pseudogene MimV: Mimiviruses, CatV: Catovirus, BsV: Bodo saltans virus, HokV: Hokovirus, KloV: Klosneuvirus (KlosnV), IndV: Indivirus, CroV: Cafeteria roenbergensis virus, PhycV: Phycodnaviridae, PanV: Pandoraviridae.
Figure 4—figure supplement 3.. Evolutionary history of translational machinery found in giant viruses inferred by COUNT and abundance of ankyrin repeat-domain genes.
The size of the black circles mapped on a cladogram of members of the Mimiviridae (see Figure 6 for full phylogenetic analysis) represents the number of gene families involved in translation at each node or tip. Blue circles indicate the number of ankyrin repeat-domain encoding genes found in each genome. Gene gain and loss events are depicted along the branches. MVc: Megavirus chilensis, AMoV: Acanthamoeba polyphaga Moumouvirus, ApMV: Acanthamoeba polyphage Mimivirus, CatV: Catovirus, BsV: Bodo saltans virus, HokV: Hokovirus, KloV: Klosneuvirus (KlosnV), IndV: Indivirus, CroV: Cafeteria roenbergensis virus, OLV1: Organic Lake Phycodnavirus 1, OLV2: Organic Lake Phycodnavirus 2, CeV: Chrysochromulina Ericina Virus, PgV: Phaeocystis globosa virus PgV-16T, AaV: Aureococcus anophagefferens virus.
Figure 5.. Invasion of RNA polymerase genes by selfish genetic elements.
Organization of self-splicing group 1 introns (grey) and inteins (black) within genes polr2a and polr2b (green). ORFs within the introns encode homing endonucleases (red). The precursor proteins (orange) are expressed after introns are excised from the pre-mRNA including the internal ORFs encoding the endonucleases. Consequently, the inteins (grey) excise themselves from the precursor protein resulting in the mature protein (yellow). Secondary structure of related self-splicing group 1 introns (‘parental’ intron polr2a-i1b and ‘offspring’ polr2a-i1a and –i1c) is shown above the coding sequence with conserved self-splicing catalytic site highlighted by arrows. Additional secondary structure predictions and sequence alignment are available in Figure 5—figure supplement 1.
Figure 5—figure supplement 1.. Self-splicing group 1 introns found in BsV.
A: DNA Sequence alignment of recently duplicated self-splicing group 1 introns polr2a-i1a and polr2a-i1c (94% DNA sequence ID). B: Secondary structure and conserved autocatalytic ribozyme centers of other polr encoded introns.
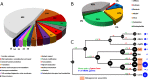
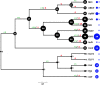
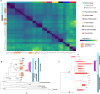
Figure 6.. BsV Phylogeny (A) Phylogenetic distance matrix of NCLDV genomes based on whole genome content of gene family presence/absence.
Both axes are identical and ‘heat’ of the color increases with dissimilarity in genome content. The shaded ‘taxa’ on the axes indicate viral sequences assembled from metagenomic data. (B) Bayesian posterior consensus tree with Bayesian posterior probability of five concatenated Nucleo-Cytoplasmic Virus Orthologous Groups (NCVOGs) from selected NCLDVs based on two independent MCMC chains (16100 generations rel_diff: 0.104001 effsize: 297). The shaded labels at the end of the branches represent ‘taxa’ based on sequences assembled from metagenomic data (C) Maximum likelihood phylogenetic tree of DNA Polymerase family B of BsV within the Mimiviridae. Branch width correlated to the distribution of 256 metagenomic sequences identified as NCLDV DNA polymerase Family B genes from the TARA oceans project recruited to the tree with pplacer.
Figure 6—figure supplement 1.. Phylogenetic analysis of five concatenated NCVOGs from selected NCLDVs.
(A) Maximum likelihood phylogenetic tree based on 1000 bootstrap replicates as calculated by RAxML (B) Maximum likelihood phylogenetic tree based on 1000 bootstrap replicates as calculated by PhyML. (C) Bayesian posterior consensus tree with Bayesian posterior probability on two independent MCMC chains (16100 generations) MVc: Megavirus chilensis, AMoV: Acanthamoeba polyphaga Moumouvirus, ApMV: Acanthamoeba polyphage Mimivirus, CatV: Catovirus, BsV: Bodo saltans virus, HokV: Hokovirus, KloV: Klosneuvirus (KlosnV), IndV: Indivirus, CroV: Cafeteria roenbergensis virus, OLV1: Organic Lake Phycodnavirus 1, OLV2: Organic Lake Phycodnavirus 2, CeV: Chrysochromulina Ericina Virus, PgV: Phaeocystis globosa virus PgV-16T, AaV: Aureococcus anophagefferens virus, FspV: Feldmannia species virus, PVd: Pandoravirus dulcis, PVs: Pandoravirus salinus, EhV: Emiliania huxleyi virus 86, PbCV: Paramecium bursaria chlorella virus 1, OlV: Ostreococcus lucimarinus virus 1, MpVS: Micromoas pusillae Virus SP-1, MpV1: Micromoas sp. RCC1109 Virus, BtV: Bathycoccus sp. RCC1105 Virus, LauV: Lausannevirus, ISKV: Infectious spleen and kidney necrosis virus, HvV: Heliothis virescens ascovirus 3e, FauV: Faustovirus E12, ASFV: African swine fever virus BA71V, VacV: Vaccinia virus, CPoV: Canary pox virus.
Figure 6—figure supplement 2.. Phylogenetic analysis of D5-like helicase-primase (NCVOG0023;A-C) and DNA polymerase family B (NCVOG0038;D-F) from selected NCLDVs.
(A/D) Maximum likelihood phylogenetic tree based on 1000 bootstrap replicates as calculated by RAxML (B/E) Maximum likelihood phylogenetic tree based on 1000 bootstrap replicates as calculated by PhyML. (C/E) Bayesian posterior consensus tree with Bayesian posterior probability on two independent MCMC chains. MVc: Megavirus chilensis, AMoV: Acanthamoeba polyphaga Moumouvirus, ApMV: Acanthamoeba polyphage Mimivirus, CatV: Catovirus, BsV: Bodo saltans virus, HokV: Hokovirus, KloV: Klosneuvirus (KlosnV), IndV: Indivirus, CroV: Cafeteria roenbergensis virus, OLV1: Organic Lake Phycodnavirus 1, OLV2: Organic Lake Phycodnavirus 2, CeV: Chrysochromulina Ericina Virus, PgV: Phaeocystis globosa virus PgV-16T, AaV: Aureococcus anophagefferens virus, PacV: Pacmanvirus, FauV: Faustovirus.
Figure 6—figure supplement 3.. Phylogenetic analysis of DNA or RNA helicases of superfamily II (NCVOG0076; A–C), packaging ATPase (NCVOG0249; D–F), Poxvirus Late Transcription Factor VLTF3-like (NCVOG0262; G–I) from selected NCLDVs.
(A/D/G) Maximum likelihood phylogenetic tree based on 1000 bootstrap replicates as calculated by RAxML (B/E/B) Maximum likelihood phylogenetic tree based on 1000 bootstrap replicates as calculated by PhyML. (C/E/I) Bayesian posterior consensus tree with Bayesian posterior probability on two independent MCMC chains. MVc: Megavirus chilensis, AMoV: Acanthamoeba polyphaga Moumouvirus, ApMV: Acanthamoeba polyphage Mimivirus, CatV: Catovirus, BsV: Bodo saltans virus, HokV: Hokovirus, KloV: Klosneuvirus (KlosnV), IndV: Indivirus, CroV: Cafeteria roenbergensis virus, OLV1: Organic Lake Phycodnavirus 1, OLV2: Organic Lake Phycodnavirus 2, CeV: Chrysochromulina Ericina Virus, PgV: Phaeocystis globosa virus PgV-16T, AaV: Aureococcus anophagefferens virus, VacV: Vaccinia virus.
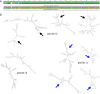
Figure 7.. Intron/intein encoded endonuclease-mediated interference completion between related viruses.
(1) Two related viruses infect the same host cell. The green virus genome contains a selfish element encoding a homing endonuclease. (2) During initial replication, the endonuclease is expressed and cleaves the unoccupied locus on the yellow virus’ genome impairing its replication. (3) Due to suppressing its competitor’s replication, the majority of the viral progeny is of the green virus’ type. (4) The yellow virus can rescue its genome by using the green virus’ genome as a template. This creates a chimeric genome containing the selfish element and the endonuclease as well as adjacent sequences originating from the green virus’ genome.
Similar articles
- Isolation of Yasminevirus, the First Member of Klosneuvirinae Isolated in Coculture with Vermamoeba vermiformis, Demonstrates an Extended Arsenal of Translational Apparatus Components.
Bajrai LH, Mougari S, Andreani J, Baptiste E, Delerce J, Raoult D, Azhar EI, La Scola B, Levasseur A. Bajrai LH, et al. J Virol. 2019 Dec 12;94(1):e01534-19. doi: 10.1128/JVI.01534-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597770 Free PMC article. - Virus Genomes from Deep Sea Sediments Expand the Ocean Megavirome and Support Independent Origins of Viral Gigantism.
Bäckström D, Yutin N, Jørgensen SL, Dharamshi J, Homa F, Zaremba-Niedwiedzka K, Spang A, Wolf YI, Koonin EV, Ettema TJG. Bäckström D, et al. mBio. 2019 Mar 5;10(2):e02497-18. doi: 10.1128/mBio.02497-18. mBio. 2019. PMID: 30837339 Free PMC article. - Hidden diversity of soil giant viruses.
Schulz F, Alteio L, Goudeau D, Ryan EM, Yu FB, Malmstrom RR, Blanchard J, Woyke T. Schulz F, et al. Nat Commun. 2018 Nov 19;9(1):4881. doi: 10.1038/s41467-018-07335-2. Nat Commun. 2018. PMID: 30451857 Free PMC article. - Updating strategies for isolating and discovering giant viruses.
Khalil JYB, Andreani J, La Scola B. Khalil JYB, et al. Curr Opin Microbiol. 2016 Jun;31:80-87. doi: 10.1016/j.mib.2016.03.004. Epub 2016 Apr 1. Curr Opin Microbiol. 2016. PMID: 27039269 Review. - Giant Viruses of Amoebae: A Journey Through Innovative Research and Paradigm Changes.
Colson P, La Scola B, Raoult D. Colson P, et al. Annu Rev Virol. 2017 Sep 29;4(1):61-85. doi: 10.1146/annurev-virology-101416-041816. Epub 2017 Jul 31. Annu Rev Virol. 2017. PMID: 28759330 Review.
Cited by
- Two classes of EF1-family translational GTPases encoded by giant viruses.
Zinoviev A, Kuroha K, Pestova TV, Hellen CUT. Zinoviev A, et al. Nucleic Acids Res. 2019 Jun 20;47(11):5761-5776. doi: 10.1093/nar/gkz296. Nucleic Acids Res. 2019. PMID: 31216040 Free PMC article. - Morphologic and Genomic Analyses of New Isolates Reveal a Second Lineage of Cedratviruses.
Rodrigues RAL, Andreani J, Andrade ACDSP, Machado TB, Abdi S, Levasseur A, Abrahão JS, La Scola B. Rodrigues RAL, et al. J Virol. 2018 Jun 13;92(13):e00372-18. doi: 10.1128/JVI.00372-18. Print 2018 Jul 1. J Virol. 2018. PMID: 29695424 Free PMC article. - A critical analysis of the current state of virus taxonomy.
Caetano-Anollés G, Claverie JM, Nasir A. Caetano-Anollés G, et al. Front Microbiol. 2023 Aug 3;14:1240993. doi: 10.3389/fmicb.2023.1240993. eCollection 2023. Front Microbiol. 2023. PMID: 37601376 Free PMC article. Review. - Characterization of Allobodo yubaba sp. nov. and Novijibodo darinka gen. et sp. nov., cultivable free-living species of the phylogenetically enigmatic kinetoplastid taxon Allobodonidae.
Packer JA, Zavadska D, Weston EJ, Eglit Y, Richter DJ, Simpson AGB. Packer JA, et al. J Eukaryot Microbiol. 2025 Jan-Feb;72(1):e13072. doi: 10.1111/jeu.13072. J Eukaryot Microbiol. 2025. PMID: 39868642 Free PMC article. - Euglenozoa: taxonomy, diversity and ecology, symbioses and viruses.
Kostygov AY, Karnkowska A, Votýpka J, Tashyreva D, Maciszewski K, Yurchenko V, Lukeš J. Kostygov AY, et al. Open Biol. 2021 Mar;11(3):200407. doi: 10.1098/rsob.200407. Epub 2021 Mar 10. Open Biol. 2021. PMID: 33715388 Free PMC article. Review.
References
- Andersen RA, Berges JA, Harrison PJ, Watanabe MM. Algal Culturing Techniques. Amsterdam: Elsevier; 2005. Recipes for freshwater and seawater media; pp. 429–538.
- Arantes TS, Rodrigues RA, Dos Santos Silva LK, Oliveira GP, de Souza HL, Khalil JY, de Oliveira DB, Torres AA, da Silva LL, Colson P, Kroon EG, da Fonseca FG, Bonjardim CA, La Scola B, Abrahão JS. The large marseillevirus explores different entry pathways by forming giant infectious vesicles. Journal of Virology. 2016;90:5246–5255. doi: 10.1128/JVI.00177-16. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous