Incidence Prediction for the 2017-2018 Influenza Season in the United States with an Evolution-informed Model - PubMed (original) (raw)
Incidence Prediction for the 2017-2018 Influenza Season in the United States with an Evolution-informed Model
Xiangjun Du et al. PLoS Curr. 2018.
Abstract
Introduction: Seasonal influenza is responsible for a high disease burden in the United States and worldwide. Predicting outbreak size in advance can contribute to the timely control of seasonal influenza by informing health care and vaccination planning.
Methods: Recently, a process-based model was developed for forecasting incidence dynamics ahead of the season, with the approach validated by several statistical criteria, including an accurate real-time prediction for the past 2016-2017 influenza season before it started.
Results: Based on this model and data up to June 2017, a forecast for the upcoming 2017-2018 influenza season is presented here, indicating an above-average, moderately severe, outbreak dominated by the H3N2 subtype.
Discussion: The prediction is consistent with surveillance data so far, which already indicate the predominance of H3N2. The forecast for the upcoming 2017-2018 influenza season reinforces the importance of the on-going vaccination campaign.
Keywords: 2017-2018; Influenza; United States; evolution; forecast; incidence; model; seasonal.
Figures
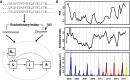
Model and Forecasts.
(A) Workflow of the EvoEpiFlu model. The evolutionary index is calculated as a weighted sum of sequence distances (based on epitope sites of HA) to compare strains each month to strains in the past, with distances weighted by a decay function back in time. Another approach relies on a discrete version of evolutionary change and is based on a previously-published genotype-phenotype map , producing the proportion of antigenic variants (PAV) over time. In order to make the forecast, two versions of the model were developed that incorporate the evolutionary index either in continuous or discrete form (in the continuous model and discrete model respectively). These quantities are included as covariates in a modified SIRS formulation for the transmission dynamics of the disease (tracking three main classes of individuals, S for susceptible, I for infected and infectious, and R for recovered and immune, as well as an additional class for immune to H1N1, which is informed by observed H1N1 incidence). In the models, evolutionary change determines the rate of return of immune individuals to the susceptible class, mimicking a loss of protection as the virus evolves. (B) Model evolutionary components and forecasts (based on available data up to November, 2017 at the time of publication). Top panel: PAV calculated for strains in each quarter compared to strains in the past 12 months. Middle panel: the evolutionary index calculated monthly. Bottom panel: the H3N2 incidence forecast based on data up to June, 2017 for the upcoming 2017-2018 season in the US based on the cluster version of EvoEpiFlu is shown in yellow. An updated forecast using data up to November 2017 is also shown in green. The observed data are indicated in black, with observed data for the 2017-2018 season up to December at the time of publication shown with the dotted line; forecasts are color-coded with their corresponding 95% uncertainty intervals, with blue for leave-one-out cross-validation, red for one-season-ahead forecasting (for out-of-fit data), and for the real-time forecast of the 2016-2017 season. For model details see , from which the forecasts up to 2016-2017 are taken.
Similar articles
- Evolution-informed forecasting of seasonal influenza A (H3N2).
Du X, King AA, Woods RJ, Pascual M. Du X, et al. Sci Transl Med. 2017 Oct 25;9(413):eaan5325. doi: 10.1126/scitranslmed.aan5325. Sci Transl Med. 2017. PMID: 29070700 Free PMC article. - Predicting the epidemic sizes of influenza A/H1N1, A/H3N2, and B: a statistical method.
Goldstein E, Cobey S, Takahashi S, Miller JC, Lipsitch M. Goldstein E, et al. PLoS Med. 2011 Jul;8(7):e1001051. doi: 10.1371/journal.pmed.1001051. Epub 2011 Jul 5. PLoS Med. 2011. PMID: 21750666 Free PMC article. - Timely vaccine strain selection and genomic surveillance improves evolutionary forecast accuracy of seasonal influenza A/H3N2.
Huddleston J, Bedford T. Huddleston J, et al. medRxiv [Preprint]. 2024 Sep 13:2024.09.11.24313489. doi: 10.1101/2024.09.11.24313489. medRxiv. 2024. PMID: 39314963 Free PMC article. Preprint. - The need for quadrivalent vaccine against seasonal influenza.
Belshe RB. Belshe RB. Vaccine. 2010 Sep 7;28 Suppl 4:D45-53. doi: 10.1016/j.vaccine.2010.08.028. Vaccine. 2010. PMID: 20713260 Review. - Recommendations for Prevention and Control of Influenza in Children, 2018-2019.
COMMITTEE ON INFECTIOUS DISEASES. COMMITTEE ON INFECTIOUS DISEASES. Pediatrics. 2018 Oct;142(4):e20182367. doi: 10.1542/peds.2018-2367. Epub 2018 Sep 3. Pediatrics. 2018. PMID: 30177511 Review.
Cited by
- Forecasting type-specific seasonal influenza after 26 weeks in the United States using influenza activities in other countries.
Choi SB, Kim J, Ahn I. Choi SB, et al. PLoS One. 2019 Nov 25;14(11):e0220423. doi: 10.1371/journal.pone.0220423. eCollection 2019. PLoS One. 2019. PMID: 31765386 Free PMC article.
References
- Luksza M, Lässig M. A predictive fitness model for influenza. Nature. 2014 Mar 6;507(7490):57-61. PubMed PMID:24572367. - PubMed
- Du X, Dong L, Lan Y, Peng Y, Wu A, Zhang Y, Huang W, Wang D, Wang M, Guo Y, Shu Y, Jiang T. Mapping of H3N2 influenza antigenic evolution in China reveals a strategy for vaccine strain recommendation. Nat Commun. 2012 Feb 28;3:709. PubMed PMID:22426230. - PubMed
Grants and funding
This work is funded by the University of Chicago. The funder had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous