Molecular imaging of glycan chains couples cell-wall polysaccharide architecture to bacterial cell morphology - PubMed (original) (raw)
Molecular imaging of glycan chains couples cell-wall polysaccharide architecture to bacterial cell morphology
Robert D Turner et al. Nat Commun. 2018.
Abstract
Biopolymer composite cell walls maintain cell shape and resist forces in plants, fungi and bacteria. Peptidoglycan, a crucial antibiotic target and immunomodulator, performs this role in bacteria. The textbook structural model of peptidoglycan is a highly ordered, crystalline material. Here we use atomic force microscopy (AFM) to image individual glycan chains in peptidoglycan from Escherichia coli in unprecedented detail. We quantify and map the extent to which chains are oriented in a similar direction (orientational order), showing it is much less ordered than previously depicted. Combining AFM with size exclusion chromatography, we reveal glycan chains up to 200 nm long. We show that altered cell shape is associated with substantial changes in peptidoglycan biophysical properties. Glycans from E. coli in its normal rod shape are long and circumferentially oriented, but when a spheroid shape is induced (chemically or genetically) glycans become short and disordered.
Conflict of interest statement
The authors declare no competing interests.
Figures
Fig. 1
Direct visualisation of glycan strand arrangement in the E. coli polymer envelope by AFM. a 3D representation of a peptidoglycan fragment mounted on poly-
l
-ornithine (see annotations). b Higher resolution image of boxed region in a. Long glycan chains are visible. c Zoom of marked boxed region in b showing side-by-side chains. d Zoom of marked boxed region in b showing overlapping chains
Fig. 2
Mapping and quantification of glycan chain arrangement. a Example AFM image data. b Gradient orientation map of a. Inset: Polar histogram of data shown in b, overlaid with fitted ellipse. Dividing the long axis of the ellipse by the short yields a dimensionless parameter which reflects orientation order, with a higher number indicating an increased tendency to order. c Location of pores identified in the example image a. Inset: distribution of pore areas. d Peptidoglycan at pole: Large scan with bacterial shape outlined (dotted line). The instability inherent in imaging overlapping polymer leaflets is apparent in the top half of the image (height range 20 nm). e Smaller scan (acquired subsequently at lower scan speed) of region marked in d (height range 4 nm). f Enlargement of region marked in e. Many glycan strands running approximately along the circumferential axis of the cell are clearly visible (height range 3.5 nm). Inset: polar histogram. g Peptidoglycan from cylinder: Large scan of a different sacculus to d (height range 7.5 nm). h Smaller scan (acquired subsequently at lower scan speed) of region marked in g (height range 4 nm). i Enlargement of region marked in h. Many glycan strands running approximately along the circumferential axis of the cell are clearly visible (height range 4 nm). Inset: polar histogram
Fig. 3
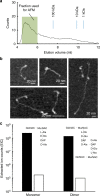
Size exclusion chromatography and AFM imaging of isolated glycan strands. a Size exclusion chromatography trace of glycan chains from E. coli (MG1655). b AFM images of glycan chains from fraction shown in a (each panel adjusted for best contrast). c Sums of extracted ion counts (EIC) for positively charged adducts of the major peptidoglycan sugar-peptide monomer and dimer from material digested with either Cellosyl alone (black bars), or ATL amidase, then Cellosyl (white bars). Amidase treatment removes the cross-linking peptides, substantially reducing the abundance of sugar-peptide adducts and freeing the glycan chains
Fig. 4
Peptidoglycan characteristics in spheroid E. coli. a Optical phase contrast microscopy image of E. coli bacterial cells grown in media containing 10 μg/ml A22. b Optical phase contrast microscopy image of E. coli bacterial cells lacking the gene encoding MreB. c Image of peptidoglycan from E. coli grown in media containing A22, and associated polar histogram. Glycan strands appear less orientationally ordered and this is reflected in the more circular shape of the polar histogram. d Image of peptidoglycan from E. coli lacking the gene encoding MreB, and associated polar histogram. Glycan strands again appear less orientationally ordered. e Size exclusion chromatography trace of glycan chains from E. coli grown in media containing 10 μg/ml A22 (black line). Chains from bacteria grown in this way are shorter than for bacteria grown in unmodified media (see trace in grey for comparison). f Size exclusion chromatography trace of glycan chains from E. coli lacking the gene encoding MreB (black line). Chains from this genetically modified bacterial strain are shorter than those of unmodified bacteria (grey line)
Fig. 5
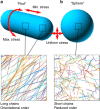
Conceptual diagrams. a Peptidoglycan from rod-shaped E. coli (MG1655) is not crystalline yet has quantifiable orientational order with chains likely to be in a circumferential direction. This corresponds to the direction of maximum stress in the cylindrical part of the cell (stress is isotropic at the poles). It contains glycans up to 200 nm long. b Peptidoglycan from roughly spherical E. coli (lacking MreB or treated with A22) is much less ordered and has shorter glycan chains. Stress is isotropic in this case
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Ceftazidime with avibactam for treating severe aerobic Gram-negative bacterial infections: technology evaluation to inform a novel subscription-style payment model.
Harnan S, Kearns B, Scope A, Schmitt L, Jankovic D, Hamilton J, Srivastava T, Hill H, Ku CC, Ren S, Rothery C, Bojke L, Sculpher M, Woods B. Harnan S, et al. Health Technol Assess. 2024 Oct;28(73):1-230. doi: 10.3310/YAPL9347. Health Technol Assess. 2024. PMID: 39487661 Free PMC article. - Dimer-monomer transition defines a hyper-thermostable peptidoglycan hydrolase mined from bacterial proteome by lysin-derived antimicrobial peptide-primed screening.
Zhang L, Hu F, Zhao Z, Li X, Zhong M, He J, Yao F, Zhang X, Mao Y, Wei H, He J, Yang H. Zhang L, et al. Elife. 2024 Nov 26;13:RP98266. doi: 10.7554/eLife.98266. Elife. 2024. PMID: 39589395 Free PMC article. - Requirement for the cell division protein DivIB in polar cell division and engulfment during sporulation in Bacillus subtilis.
Thompson LS, Beech PL, Real G, Henriques AO, Harry EJ. Thompson LS, et al. J Bacteriol. 2006 Nov;188(21):7677-85. doi: 10.1128/JB.01072-06. Epub 2006 Aug 25. J Bacteriol. 2006. PMID: 16936026 Free PMC article. - Metformin for endometrial hyperplasia.
Shiwani H, Clement NS, Daniels JP, Atiomo W. Shiwani H, et al. Cochrane Database Syst Rev. 2024 May 2;5(5):CD012214. doi: 10.1002/14651858.CD012214.pub3. Cochrane Database Syst Rev. 2024. PMID: 38695827 Review.
Cited by
- AFM-Based Correlative Microscopy Illuminates Human Pathogens.
Bhat SV, Price JDW, Dahms TES. Bhat SV, et al. Front Cell Infect Microbiol. 2021 May 7;11:655501. doi: 10.3389/fcimb.2021.655501. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34026660 Free PMC article. Review. - Simulations suggest a constrictive force is required for Gram-negative bacterial cell division.
Nguyen LT, Oikonomou CM, Ding HJ, Kaplan M, Yao Q, Chang YW, Beeby M, Jensen GJ. Nguyen LT, et al. Nat Commun. 2019 Mar 19;10(1):1259. doi: 10.1038/s41467-019-09264-0. Nat Commun. 2019. PMID: 30890709 Free PMC article. - Allosteric activation of cell wall synthesis during bacterial growth.
Shlosman I, Fivenson EM, Gilman MSA, Sisley TA, Walker S, Bernhardt TG, Kruse AC, Loparo JJ. Shlosman I, et al. Nat Commun. 2023 Jun 10;14(1):3439. doi: 10.1038/s41467-023-39037-9. Nat Commun. 2023. PMID: 37301887 Free PMC article. - The hidden base of the iceberg: gut peptidoglycome dynamics is foundational to its influence on the host.
Wheeler R, Gomperts Boneca I. Wheeler R, et al. Gut Microbes. 2024 Jan-Dec;16(1):2395099. doi: 10.1080/19490976.2024.2395099. Epub 2024 Sep 6. Gut Microbes. 2024. PMID: 39239828 Free PMC article. Review. - An Updated Model of the Divisome: Regulation of the Septal Peptidoglycan Synthesis Machinery by the Divisome.
Attaibi M, den Blaauwen T. Attaibi M, et al. Int J Mol Sci. 2022 Mar 24;23(7):3537. doi: 10.3390/ijms23073537. Int J Mol Sci. 2022. PMID: 35408901 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous