TISSUES 2.0: an integrative web resource on mammalian tissue expression - PubMed (original) (raw)
TISSUES 2.0: an integrative web resource on mammalian tissue expression
Oana Palasca et al. Database (Oxford). 2018.
Erratum in
- TISSUES 2.0: an integrative web resource on mammalian tissue expression.
Palasca O, Santos A, Stolte C, Gorodkin J, Jensen LJ. Palasca O, et al. Database (Oxford). 2018 Jan 1;2018:bay028. doi: 10.1093/database/bay028. Database (Oxford). 2018. PMID: 30403794 Free PMC article. No abstract available.
Abstract
Abstract: Physiological and molecular similarities between organisms make it possible to translate findings from simpler experimental systems—model organisms—into more complex ones, such as human. This translation facilitates the understanding of biological processes under normal or disease conditions. Researchers aiming to identify the similarities and differences between organisms at the molecular level need resources collecting multi-organism tissue expression data. We have developed a database of gene–tissue associations in human, mouse, rat and pig by integrating multiple sources of evidence: transcriptomics covering all four species and proteomics (human only), manually curated and mined from the scientific literature. Through a scoring scheme, these associations are made comparable across all sources of evidence and across organisms. Furthermore, the scoring produces a confidence score assigned to each of the associations. The TISSUES database (version 2.0) is publicly accessible through a user-friendly web interface and as part of the STRING app for Cytoscape. In addition, we analyzed the agreement between datasets, across and within organisms, and identified that the agreement is mainly affected by the quality of the datasets rather than by the technologies used or organisms compared.
Database url: http://tissues.jensenlab.org/
Figures
Figure 1.
Schematic representation for calculating fold enrichment and fitting its relationship to raw expression values. For a given dataset, we select gene–tissue pairs and their raw expression scores for a subset of tissues, which are common between all datasets and a subset of genes common between the dataset and the gold standard. Next, we sort these gene–tissue pairs by their raw expression value, and traverse them in sliding windows of a pre-defined size. The enrichment corresponding to each bin is then calculated as the fraction of gene–tissue pairs from that bin found in the gold standard, divided by the fraction of pairs that would be expected by random. Next, we use appropriate functions (see text) to fit the relationship between fold enrichment values and mean raw expression values in their corresponding windows.
Figure 2.
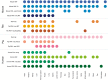
Summary of tissues present in each dataset. We mapped the newly integrated datasets from mouse, rat and pig, as well as the already existing human datasets, to 21 major tissues of interest. This figure shows which of these tissues are covered by which datasets.
Figure 3.
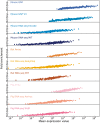
In order to obtain confidence scores for each gene–tissue pair, we assess the relationship between raw expression values and fold enrichment, defined as the agreement between each dataset and gold standard datasets specific to the organism. The gold standard datasets are based on the UniProtKB protein tissue annotations in human, filtered for 1-to-1 orthologs between human and each of the three organisms. The _x_-axis contains raw expression values for gene–tissue pairs, in units specific to the type of experiment or processing of data (e.g. intensity units for microarray studies, FPKMs for RNA-seq), and averaged across bins of 100 pairs.
Figure 4.
(A) Pearson’s correlation coefficients between final confidence scores of gene–tissue associations across datasets. For each pair of datasets, we considered the set of common genes (genes being expressed in at least one tissue in each dataset), and common tissues between the two datasets. (B) In this panel, we compare the rates of True positives and False positives for each dataset for the common tissues to show that the correlation between datasets is mainly influenced by quality rather than by organism or technology.
Figure 5.
Summary figures for all the covered organisms. The web interface provides a comprehensive figure for each organism where the tissue associations for the queried gene are summarized. In this example, we are showing the tissue expression profile for the Microtubule-associated protein tau (MAPT) known to be related to Alzheimer’s disease (57). The ortholog–paralog table provides information about homologous proteins and their tissue–expression correlation with the query protein.
Figure 6.
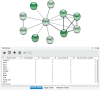
STRING app in Cytoscape with tissue information. (A) The newly developed stringApp for Cytoscape allows users to get all the STRING functionality within Cytoscape and allows expression evidence from TISSUES to be visualized onto the network (in this example for liver). (B) The stringApp also shows the evidence score for each of the major tissues in the node attributes table.
Similar articles
- MSGD: a manually curated database of genomic, transcriptomic, proteomic and drug information for multiple sclerosis.
Wu T, Hou Y, Xin G, Niu J, Peng S, Xu F, Li Y, Chen Y, Yu Y, Zhang H, Kong X, Cao Y, Ning S, Wang L, Hao J. Wu T, et al. Database (Oxford). 2024 May 24;2024:baae037. doi: 10.1093/database/baae037. Database (Oxford). 2024. PMID: 38788333 Free PMC article. - SCPortalen: human and mouse single-cell centric database.
Abugessaisa I, Noguchi S, Böttcher M, Hasegawa A, Kouno T, Kato S, Tada Y, Ura H, Abe K, Shin JW, Plessy C, Carninci P, Kasukawa T. Abugessaisa I, et al. Nucleic Acids Res. 2018 Jan 4;46(D1):D781-D787. doi: 10.1093/nar/gkx949. Nucleic Acids Res. 2018. PMID: 29045713 Free PMC article. - RAIN: RNA-protein Association and Interaction Networks.
Junge A, Refsgaard JC, Garde C, Pan X, Santos A, Alkan F, Anthon C, von Mering C, Workman CT, Jensen LJ, Gorodkin J. Junge A, et al. Database (Oxford). 2017 Jan 10;2017:baw167. doi: 10.1093/database/baw167. Print 2017. Database (Oxford). 2017. PMID: 28077569 Free PMC article. - Rat Genome Database (RGD): mapping disease onto the genome.
Twigger S, Lu J, Shimoyama M, Chen D, Pasko D, Long H, Ginster J, Chen CF, Nigam R, Kwitek A, Eppig J, Maltais L, Maglott D, Schuler G, Jacob H, Tonellato PJ. Twigger S, et al. Nucleic Acids Res. 2002 Jan 1;30(1):125-8. doi: 10.1093/nar/30.1.125. Nucleic Acids Res. 2002. PMID: 11752273 Free PMC article. - NeuroTransDB: highly curated and structured transcriptomic metadata for neurodegenerative diseases.
Bagewadi S, Adhikari S, Dhrangadhariya A, Irin AK, Ebeling C, Namasivayam AA, Page M, Hofmann-Apitius M, Senger P. Bagewadi S, et al. Database (Oxford). 2015 Oct 16;2015:bav099. doi: 10.1093/database/bav099. Print 2015. Database (Oxford). 2015. PMID: 26475471 Free PMC article.
Cited by
- COVID-19 and Alzheimer's disease: how one crisis worsens the other.
Xia X, Wang Y, Zheng J. Xia X, et al. Transl Neurodegener. 2021 Apr 30;10(1):15. doi: 10.1186/s40035-021-00237-2. Transl Neurodegener. 2021. PMID: 33941272 Free PMC article. Review. - Autoimmune Encephalitis in COVID-19 Infection: Our Experience and Systematic Review of the Literature.
Stoian A, Stoian M, Bajko Z, Maier S, Andone S, Cioflinc RA, Motataianu A, Barcutean L, Balasa R. Stoian A, et al. Biomedicines. 2022 Mar 25;10(4):774. doi: 10.3390/biomedicines10040774. Biomedicines. 2022. PMID: 35453524 Free PMC article. Review. - In silico prediction of HIV-1-host molecular interactions and their directionality.
Chai H, Gu Q, Hughes J, Robertson DL. Chai H, et al. PLoS Comput Biol. 2022 Feb 8;18(2):e1009720. doi: 10.1371/journal.pcbi.1009720. eCollection 2022 Feb. PLoS Comput Biol. 2022. PMID: 35134057 Free PMC article. - Viruses.STRING: A Virus-Host Protein-Protein Interaction Database.
Cook HV, Doncheva NT, Szklarczyk D, von Mering C, Jensen LJ. Cook HV, et al. Viruses. 2018 Sep 23;10(10):519. doi: 10.3390/v10100519. Viruses. 2018. PMID: 30249048 Free PMC article. - Identification of HOX signatures contributing to oral cancer phenotype.
Padam KSR, Morgan R, Hunter K, Chakrabarty S, Kumar NAN, Radhakrishnan R. Padam KSR, et al. Sci Rep. 2022 Jun 16;12(1):10123. doi: 10.1038/s41598-022-14412-6. Sci Rep. 2022. PMID: 35710803 Free PMC article.
References
- Aitman T.J., Boone C., Churchill G.A., et al. (2011) The future of model organisms in human disease research. Nat. Rev. Genet., 12, 575–582. - PubMed
- Greaves P., Williams A., Eve M. (2004) First dose of potential new medicines to humans: how animals help. Nat. Rev. Drug Disc., 3, 226–236. - PubMed
- Boverhof D.R., Chamberlain M.P., Elcombe C.R.. et al. (2011) Transgenic animal models in toxicology: historical perspectives and future outlook. Toxicol. Sci., 121, 207–233. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources